ECB-FEAT-23045511
Strongylocentrotus purpuratus Genome Assembly v5.0
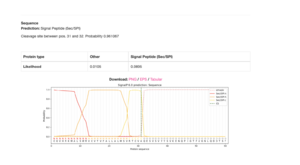
The gene model for LOC580282 appears to be incomplete, with the upstream locus, LOC115922232, most likely being part of this gene based on several lines of evidence. The lines of evidence for a split-gene misannotation include the lack of signal peptide sequences yielded by SMART (Letunic et al., 2006) and SignalP – 6.0 analyses (Teufel et al., 2022), the absence of a 5’-UTR in the mRNA transcript, XM_030981285.1, and the adjacency of the coding loci on scaffold NW_022145596.1.



Lytechinus variegatus Genome Assembly v3.0
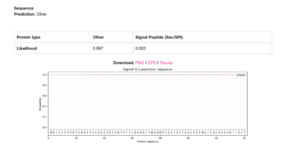
The gene model, LOC121430458, appears to be incomplete, with the upstream locus, LOC121412825, actually being part of the gene based on several lines of evidence. These lines of evidence include the lack of signal peptide sequences yielded by SMART (Letunic et al., 2006) and Signal 6.0P analyses (Teufel et al., 2022), the absence of a 5’-UTR of the mRNA transcript (XM_041627728.1), and the adjacency of the coding loci on chr1.