|
|
| Line 1: |
Line 1: |
| | <span class="newwin">[http://new.echinobase.org/literature/article.do?method=display&articleId=42133 '''Sea Urchin Fertilization Laboratory''']</span><br /> |
| | Vic Vacquier developed a handout for university students on how to observe sea urchin fertilization in a university lab class setting. All that is needed are sea urchin gametes, a microscope, a few solutions and this handout. The handout works well for both 1 or 2 three hour labs. The student reads through the handout, makes the simple observations, and does the simple, direct experiments to gain insight into the fascinating process of sperm-egg interaction and egg activation. |
|
| |
|
| Welcome to the Echinobase CRISPR/Cas resource. A brief literature and method review is followed by tables of gRNA spacer sequences.
| |
|
| |
|
| Updated December 2020
| | <span class="newwin">'''Ocean Life Class for K-5'''</span><br /> |
|
| |
|
| | <p>Class objective is to instill a sense of wonder and curiosity at the diversity of invertebrate life and forms in the ocean.</p> |
| | <p>The worksheets that follow are suggested to be used in a class involving live marine invertebrates purchased from marine animal suppliers.</p> |
| | <p>The discussions and activities depend upon the facilities available and the area of study of the host aquarium and lab.</p> |
|
| |
|
| '''''S. purpuratus'' genome editing to create insertions and deletions'''
| |
|
| |
|
| To date CRISPR/Cas9 has been used to introduce insertion and deletion mutations (indels) into ''S. purpuratus nodall'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016]), ''polyketide synthase 1'', ''gcml'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016]), ''nanos2l'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=45207 Oulhen et al. 2017]) and ''dll1'' (''delta'') ([https://www.echinobase.org/literature/article.do?method=display&articleId=45720 Mellott et al. 2017]) genes. Attempts to mutate ''foxy'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=47178 Oulhen et al. 2019]) were unsuccessful. A number of different methods were used for gRNA synthesis (several using pT7-gRNA) and NLS-SpCas9-NLS (pCS2-nCas9n (zebrafish codon-optimized), or pCS2-3xFLAG-NLS-SpCas9-NLS (codon optimized for human with a 3XFLAG-tag) were used in these studies (see below for details). The gRNAs and mRNAs were microinjected into fertilized eggs.
| | <span class="newwin">'''Ocean Life Introduction'''</span><br /> |
|
| |
|
| | The oceans are rich with life, this sheet shows common marine invertebrates. |
| | <p style="margin-left: 20px">[[File:Ocean_Life.jpg|frameless|200px]]</p> |
|
| |
|
| '''Single nucleotide edits'''
| |
|
| |
|
| Additional studies fused a deaminase to two mutants of SpCas9 for achieving targeted, single nucleotide edits to ''[https://www.echinobase.org/gene/showgene.do?method=display&geneId=23094247& alx1]'', ''[https://www.echinobase.org/gene/showgene.do?method=display&geneId=23083023 segment polarity protein dishevelled homolog DVL-3]'' (''Dsh'') and ''[https://www.echinobase.org/gene/showgene.do?method=display&geneId=23139056 polyketide synthase 1]'' (''Pks1'') to produce STOP codons ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017]).
| | <span class="newwin">'''Ocean Life Word Search'''</span><br /> |
|
| |
|
| | Word searches are fun for students of all ages. |
| | [https://puzzlemaker.discoveryeducation.com/word-search Here is a site] that can be used to make puzzles and a list of terms to get you started.<br /> |
| | <p style="margin-left: 20px">MARINE</p> |
| | <p style="margin-left: 20px">ENVIRONMENT</p> |
| | <p style="margin-left: 20px">INVERTEBRATES</p> |
| | <p style="margin-left: 20px">ECHINODERMS</p> |
| | <p style="margin-left: 20px">SEAURCHIN</p> |
| | <p style="margin-left: 20px">SEASTAR</p> |
| | <p style="margin-left: 20px">ANEMONE</p> |
| | <p style="margin-left: 20px">CORAL</p> |
| | <p style="margin-left: 20px">SPONGES</p> |
| | <p style="margin-left: 20px">FISH</p> |
| | <p style="margin-left: 20px">ECHINOBASE</p> |
| | <p style="margin-left: 20px">DEVELOPMENT</p> |
| | <p style="margin-left: 20px">LARVAE</p> |
| | <p style="margin-left: 20px">MEASUREMENT</p><br /> |
|
| |
|
| '''Reviews''' | | <span class="newwin">'''Echinoderm Worksheet'''</span><br /> |
|
| |
|
| Reviews of the methods are available ([https://www.echinobase.org/literature/article.do?method=display&articleId=45567 Cui et al. 2017]; [https://www.echinobase.org/literature/article.do?method=display&articleId=47096 Lin et al. 2019]).
| | This worksheet can be used as an introduction to echinoderms.<br /> |
|
| |
|
| | <p style="margin-left: 20px">[[File:Echinoderm_Worksheet.jpg|frameless|200px]]</p> |
|
| |
|
| '''Editing other echinoderm species'''
| |
|
| |
|
| Editing technology has also been used in ''Hemicentrotus pulcherrimus'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=47348 Liu et al. 2019]; [https://www.echinobase.org/literature/article.do?method=display&articleId=48597 Wessel et al. 2020]) and ''Temnopleurus reevesii'' ([https://www.echinobase.org/literature/article.do?method=display&articleId=48853 Yaguchi et al. 2020]).
| | <span class="newwin">'''Cut and Paste Echinoderm Life Cycles'''</span><br /> |
|
| |
|
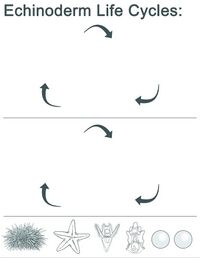
| | This image shows the life cycle of the bat star.<br /> |
|
| |
|
| '''Design overview'''
| | <p style="margin-left: 20px">[[File:Sea_Star_Life_Cycle.jpg|frameless|200px]]</p> |
|
| |
|
| CRISPR systems in nature are composed of the Cas9 nuclease and two RNAs, the CRISPR RNA (crRNA) that binds to a complementary DNA sequence and binds to the transactivating RNA (tracrRNA) that also binds to a specific Cas9 protein. For ease of use the crRNA and tracrRNA have been combined into a single guide RNA (sgRNA) molecule for use with the ''Streptococcus pyogene''s Cas9 (SpCas9). The sgRNA has the target gene '''spacer''' sequence and the '''scaffold''' sequence that interacts with the Cas9 protein. The design of the gRNA target gene specific spacer sequence can be performed using online tools such as CRISPRscan. Briefly the software will scan for the NGG protospacer adjacent motif (PAM) sequence then evaluate the 20 nucleotides that are 5' of the PAM site for their suitability as a spacer sequence. Favorable spacer sequences are more than 50% GC, 20 nucleotides in length and do not have "off-target" binding sites. Additionally, if using T7 RNA polymerase to produce the sgRNA then two '''5' GGs''' should be considered, editing occurred with an 80% frequency with GG-, 75% NG-, 60% GN- and 37.5% if NN- ([https://www.echinobase.org/literature/article.do?method=display&articleId=48856 Thomas et al. 2014]).
| |
|
| |
|
| | The students then cut and paste the bat star and then the sea urchin life cycle stages noting similarities and differences.<br /> |
|
| |
|
| '''Method overview'''
| | <p style="margin-left: 20px">[[File:Echinoderm_Life_Cycles.jpg|frameless|200px]]</p> |
|
| |
|
| Published methods have used microinjection of RNA into embryos.
| |
|
| |
|
| The capped mRNA for ''Streptococcus pyogenes'' Cas9 with nuclear localization signals was produced using either linearized [https://www.addgene.org/47929/ pCS2-nCas9n] or [https://www.addgene.org/51307/ pCS2-3XFLAG-NLS-SpCas9-NLS] as the template for the MEGAscript SP6 Transcription Kit or the mMESSAGE mMACHINE SP6 Transcription Kit. The RNA was then purified.
| |
|
| |
|
| To make the gRNAs several approaches have been used. The [https://www.addgene.org/46759/ pT7-gRNA] was designed to clone the gene specific spacer/target sequence into BsmBI restriction sites. The vector contains the T7 promoter and the gRNA scaffold followed by a restriction site for linearization prior to RNA production. More recently the pT7-gRNA plasmid has been used as template for a primer containing the T7 promoter, spacer sequence and an overlap sequence to prime the PCR and add the scaffold. This overlap approach has also been used with a synthesized scaffold oligo for PCR (eg. 5’ AAAAGCACCG ACTCGGTGCC ACTTTTTCAA GTTGATAACG GACTAGCCTT ATTTTAACTT GCTATT<u>TCTA GCTCTAAAAC</u> 3' where the overlap sequence is underlined) . The sgRNA is then produced using the MEGAshortscript T7 Transcription Kit and RNA is purified.
| | <span class="newwin">'''Morphology Activity'''</span><br /> |
|
| |
|
| For microinjection the 500-1000 ng/ul Cas9 mRNA and 150-400 ng/ul sgRNA are mixed (literature varies). The NLS-Cas9-NLS protein is approximately 4.4X the mass of gRNAs so sgRNAs are in excess. If 50pl is injected this is on the order of 10^7 molecules of Cas9 mRNA and 10^8 molecules of sgRNA.
| | This activity involves a worksheet for scoring morphology and then a cut and paste activity for scoring attributes. |
|
| |
|
| | <p style="margin-left: 20px">[[File:Body_Parts.jpg|frameless|200px]]</p> |
| | <p style="margin-left: 20px">[[File:Morphology_Activity_Page_6.jpg|frameless|200px]]</p> |
| | <p style="margin-left: 20px">[[File:Morphology_Activity_Page_2.jpg|frameless|200px]]</p> |
| | <p style="margin-left: 20px">[[File:Morphology_Activity_Page_3.jpg|frameless|200px]]</p> |
| | <p style="margin-left: 20px">[[File:Morphology_Activity_Page_4.jpg|frameless|200px]]</p> |
| | <p style="margin-left: 20px">[[File:Morphology_Activity_Page_5.jpg|frameless|200px]]</p> |
|
| |
|
| '''Table of sgRNA sequences.'''
| | <br/> |
| This table shows the gene specific spacer RNA sequence that is 5' to the constant scaffold RNA sequence. The gRNA binds to the reverse complement of the DNA sequence indicated (other strand).
| |
| | |
| {| style="border:solid 1px black" class="sortable wikitable"
| |
| !Name !!Synonym !!sgRNA sequence 5-3'!!DNA sequence!!Edits Yes/NO!!References
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E1_123
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCCUCAGGUCGCACCAGA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCCTCAGGTCGCACCAGA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| NO
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E1_157
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGCGUCCGGUGUGAUAAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGCGTCCGGTGTGATAAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E1_272
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAAGAAUGCCAAGCCAUUG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAAGAATGCCAAGCCATTG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA4
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E2_63
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGACAGCUUCGUUGGCGGGC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGACAGCTTCGTTGGCGGGC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA5
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E2_777
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGUAGGCGUUGAACUGCUUU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTAGGCGTTGAACTGCTTT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nodall gRNA6
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| E2_885
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGUGUCCGUCUCUCGGGU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTGTCCGTCTCTCGGGT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=44372 Lin and Su 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.PKS1.175(+)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGUGGUGUCUUUGUCGGUAU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GTGGTGTCTTTGTCGGTAT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.PKS1.547(-)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGUGAGGGGUUUGAGGACAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| TGAGGGGTTTGAGGACAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.PKS1.806(-)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAGAGGGUUGUCUUUGGUG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GAGAGGGTTGTCTTTGGTG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| gcml gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.GCM.63(+)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCCGCCGGAGCUGCCGGGU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GCCGCCGGAGCTGCCGGGT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| | |
| |
| |
| <div class="style8" align="left">
| |
| | |
| gcml gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.GCM.390(+)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCUGUUCGGCCAGCCACUU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| CTGTTCGGCCAGCCACTT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| gcml gRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.GCM.541(+)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGAUCCUAUUCCAAUCGAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GATCCTATTCCAATCGAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| NO
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| gcml gRNA4
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp.GCM.944(-)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGAGUCCAGCCGUCCAUCU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GAGTCCAGCCGTCCATCT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=48855 Oulhen and Wessel 2016])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nanos2l gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp nanos2.190
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGUGACUGGCUCGUCGAGAC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| TGACTGGCTCGTCGAGAC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45207 Oulhen et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nanos2l gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp nanos2.250
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGAUCUCAGCGAUGUUCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GATCTCAGCGATGTTCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| NO
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45207 Oulhen et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nanos2l gRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp nanos2.295
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAGGAAGGCGAGCCAACAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAGGAAGGCGAGCCAACAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45207 Oulhen et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| nanos2l gRNA4
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Sp nanos2.319
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAGGUGGUGCUACGGGUGU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GAGGTGGTGCTACGGGTGT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45207 Oulhen et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| dll gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Delta-1 (19D10)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGUGAACUGGUAGCACGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGTGAACTGGTAGCACGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45720 Mellott et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| dll gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Delta-2 (20D11)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCUACACGUGCCUCUGUCC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCTACACGTGCCTCTGTCC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45720 Mellott et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| dll gRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Delta-3 (20D14)
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGACCGAAUCAGAUUCCGCG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGACCGAATCAGATTCCGCG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| nd
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45720 Mellott et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| alx1 gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Alx1-sgRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGAGAACAGGCGCGCCAAGUGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| AGAACAGGCGCGCCAAGTG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| alx1 gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Alx1-sgRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCGUGGGGGGCCUCAACCCGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GCGTGGGGGGCCTCAACCCGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC584189 gRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Dsh-sgRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGUAGCUUAGCCAGGCCGAUG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTAGCTTAGCCAGGCCGATG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC584189 gRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Dsh-sgRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCAUCGGUCCCCCUAGCCAGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGCATCGGTCCCCCTAGCCAGG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA4
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA1
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGAGGUUCUCAAGGAAGGUA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGAGGTTCTCAAGGAAGGTA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA5
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA2
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGUUCUUUAAGAUCUCGCC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GTTCTTTAAGATCTCGCC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA6
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA3
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGUGGUGUCUUUGUCGGUAU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GTGGTGTCTTTGTCGGTAT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA7
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA4
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGUAUUGCGCACAGUGUGU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTATTGCGCACAGTGTGT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA8
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA5
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGCGACACAGCUUGUGCCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| CGACACAGCTTGTGCCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA9
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA6
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGUUGUCCUCAAACCCCUCA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| TTGTCCTCAAACCCCTCA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA10
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA7
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGUAGCGCCAUCGCAGCCAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTAGCGCCATCGCAGCCAA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA11
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA8
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGCACUAUGUCGAAGCUCA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GCACTATGTCGAAGCTC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA12
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA9
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGUGAUCCUCUGGAAGCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGTGATCCTCTGGAAGCAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA13
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA10
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGCACCAAAGACAACCCUCUC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GCACCAAAGACAACCCTCTC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA14
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA11
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGCAACUUUGGACAUACCG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GCAACTTTGGACATACCG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA15
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA12
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGUACCAUCCCACCAACCAU
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GTACCATCCCACCAACCAT
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA16
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA13
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGAAGACAAGCACAUCAUUG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GAAGACAAGCACATCATTG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA17
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA14
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGAGGAGCUGACUCCAGAAC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| AGGAGCTGACTCCAGAAC
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA18
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA15
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGACAAGGAAGGACAGCCUA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GACAAGGAAGGACAGCCTA
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div>
| |
| |-
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| LOC588806 gRNA19
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| Pks-sgRNA16
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GGGGGACAGCUGUUGCCAAACAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| GACAGCTGTTGCCAAACAG
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="center">
| |
| | |
| Yes
| |
| | |
| </div>
| |
| |
| |
| <div class="style8" align="left">
| |
| | |
| ([https://www.echinobase.org/literature/article.do?method=display&articleId=45725 Shevidi et al. 2017])
| |
| | |
| </div> | |
| |-
| |