imported>Ctelmer |
imported>BArsh |
| Line 1: |
Line 1: |
| <p> The Davidson laboratory at Caltech generated a panel of quantitative PCR primers useful for measuring mRNA abundance of genes involved in early development, particularly those in the endomesoderm gene regulatory network. Below is the table of primer sequences.</p>
| | {| |
| | | |[[File:seaurchin.png|thumb|Adult sea urchin © Ann Cutting, California Institute of Technology]] |
| <p>Note that the gene names listed in the table have been updated according to nomenclature guidelines and may have changed on the gene pages in Echinobase, however, they are listed as synonyms and the gene pages can be found by searching for the name listed in the table.</p>
| | |[[File:Spurp_3323.jpg|thumb|Strongylocentrotus purpuratus 4 cell stage embryo. Two-cell injection of lineage tracers. © Andy Cameron, California Institute of Technology]] |
| | | |[[File:ACutting4.jpg|thumb|Strongylocentrotus purpuratus Sea Urchin: Ann Cutting, California Institute of Technology ©2014]] |
| | | |[[File:ACutting5.jpg|thumb|Strongylocentrotus purpuratus Sea Urchin: Ann Cutting, California Institute of Technology ©2014]] |
| | |
| == qPCR Primer Table ==
| |
| | |
| | |
| {| style="border:solid 1px black" class="sortable"
| |
| !GENE!!Full Name!!Contributor!!Primer seq (Forward)!!Primer seq (Reverse)!!Primer Length!!Tm!!Product size!!Comments from Q-PCR!!Source for seq. info
| |
| |- | | |- |
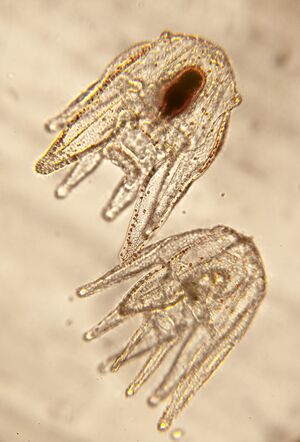
| |18S||18 S ribosomal RNA||AR||CAGGGTTCGATTCCGTAGAG||CCTCCAGTGGATCCTCGTTA||20/20||59.7/60.1||185 bp||works well; clean disassociation curve||Turbeville et al 1994; NCBI: L28055 | | |[[File:ACutting1.jpg|thumb|Strongylocentrotus purpuratus larva Ann Cutting, California Institute of Technology ©2014]] |
| | |[[File:ACutting3.jpg|thumb|Strongylocentrotus purpuratus larva Ann Cutting, California Institute of Technology ©2014]] |
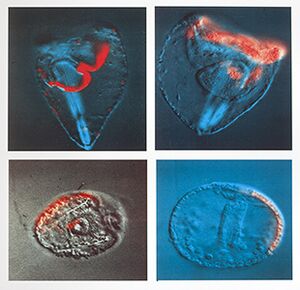
| | |[[File:LineageTracingDifferentCellsSpurp_3318.jpg|thumb|Various images of embryonic cell lineage tracings using fluorescent dye markers. © Andy Cameron, California Institute of Technology]] |
| | |[[File:ACutting2.jpg|thumb|Strongylocentrotus purpuratus larva: Ann Cutting,California Institute of Technology ©2014]] |
| |- | | |- |
| |Alx||||PO||CAGTGCAGCTTTACGTGGAC||TTAAGTCTCGGCACGACAAA|||||||||| | | |[[File:SpurpLineageTracing2color_332|thumb|Strongylocentrotus purpuratus two color lineage tracing © Andy Cameron, California Institute of Technology]] |
| |-
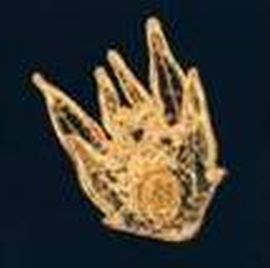
| | |[[File:golden_sea_urchin.jpg|thumb|Strongylocentrotus purpuratus © Andy Cameron, California Institute of Technology]] |
| |Apo L||Apolipophorin||JR||AGAAGAGCATCGTGCAATGA||CAGCCATGGTGTTAGCAATG||20/20||59.55/60.13||154||works well; clean disassociation curve||Cdna
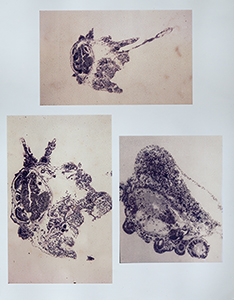
| | |[[File:Spurp_sections_3337.jpg|thumb|Histological sections of Strongylocentrotus purpuratus larvae undergoing metamorphosis © Andy Cameron, California Institute of Technology]] |
| |-
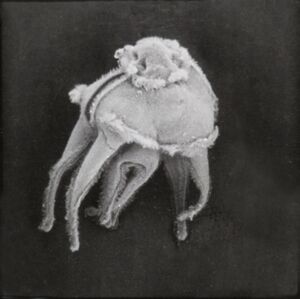
| | |[[File:Spurp_3301.jpg|thumb|SEM image of Strongylocentrotus purpuratus larva © Andy Cameron, California Institute of Technology]] |
| |APOBEC||cytidine deaminase||JR||ACCCAGTTTCACCCTCCTCT||AGGCACTCAGCTGCAAAGTT||20/20||59.97/60.20||162||ok||Bra screen
| | |
| |-
| |
| |Blimp1a||SpKrox||JS||GACCGAGGTCGATTACCAGA||CCGCGTACCTTTTGGTATGT||20/20||||175||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Blimp1b||Spkrox early form||JS||TCGCTATGCGGGATCTCTAC||GGGGTCCTTGACCTCGTAA||20/19||56.1/56.3||206||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |BMP2/4||Ligand||JN||CCAGCAAGGTCGAAGAACTC||CTCTACCCGACGACGATGAT||20/20||~60/~60||126bp||works well; clean disassociation curve||
| |
| |-
| |
| |BRA||Brachyury||CL||ACACATCGACCCATCATCAA||CATGGTGTCGTATCTTGGAAAG||20/22||59.77/59.49||139||works well; clean disassociation curve||
| |
| |-
| |
| |Brn1-2-4 (UI)||Brn1-2-4 (novel POU domain transcription factor)||CY||GTCGCATTAAGCTCGGCTAC||CAGCGGCTTCAGTTTACACA||||||||||
| |
| |-
| |
| |CAPK||SMC||JR||ccaagtacgcaggaggaaga||gagagcatcggctattgtca||20/20||60.39/58.98||100bp||OK||Endoderm/Ectoderm diff. Scr. Larval Cdna
| |
| |-
| |
| |CAT||CAT REPORTER||||TCCGGCCTTTATTCACATTC||CGGTGTAACAAGGGTGAACA||20/20||53.9/55.1||||||
| |
| |-
| |
| |Cyclophillin||PMCspecific||GA||CCAAAGTGATGGAGGTGCTT||ACAATCGTGTATGGGCAAAT||20/20||60.1/57.8||161 bp||works well, clean dissociation curve||cDNA from 40h lyb
| |
| |-
| |
| |CyIIIa||||PO||ACGCTGCAGGATTTGTAATG||TGCCAATGACAATCCCTATG||20/20||58.80/59.36||146BP||not tested, ordered||
| |
| |-
| |
| |cyIIIa - UTR||cyIIIa mRNA, against 3' UTR||TB||GAAGAACAAAAATAAAACGCATCTG||ATTGTTTGTATTGCATTCTCCAATC||25/25||60/60||60||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |cyIIIa int1||cyIIIa intron probe||TB||TGAACAAAACTGTGAAATGTGAAA||GGGCAGGGATAAAGTACCATC||24/21||60/60||108||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Dec||Decorin||CC||TTTATCTAGTTACACTTTTGCTGGTGA||ATAGATTGAGATTATCAAAGCCTCCT||27/26||59.7/59.1||300bp||works well, clean dissociation curve||cDNA from 20 hrs arrayed lib.
| |
| |-
| |
| |Dec II||Decorin||CC||GTCCAGGTGGAACACCAGAT||TGCTCCTAATGGTACGTTGACA||20/22||59.82/60.55||158bp||works well, clean dissociation curve||cDNA from 20 hrs arrayed lib.
| |
| |-
| |
| |DELTA||||PO||ACGGAGCTACATGCCTGAAC||TCACAATGGACCGAATCAGA||20/20||60.29/60.05||151bp||works well, clean dissociation curve||H.Sweet cDNA
| |
| |-
| |
| |Dop T||Dopamine tautomerase||JR||CGAGTTCGCGTACAGCATAG||GAATCCTTCGGGAAACTGCT||20/20||59.66/60.58||143||works well; clean disassociation curve||Cdna
| |
| |-
| |
| |Dri||PMCs 15-20h; Oral Ectod after25h||GA||GGTTTCCCTAGGCAAGGAAC||GACAAGATGCTGCTGTTGGA||20/20||58.9/59.4||147bp||works well, clean dissociation curve||cDNA from 40harrayed lyb
| |
| |-
| |
| |E(spl)-1||Enhancer of split-1||CC||AGTCACAGTTCCCCAGCAAC||GTGACTGTGGTGGTGTCAGG||20/20||60.16/60.05||200||works well, clean dissociation curve||partial seq from Posakony's lab
| |
| |-
| |
| |ECM3||ECM protein (see Ettensohn)||AR||AGGGCAGTGATGTTGCCTAC||GTTTGCAGCAGGGTCGTATT||20/20||60.1/60.1||161 bp||works well; clean disassociation curve||cDNA clone from 12 hr arrayed lib.
| |
| |-
| |
| |Ef1||Elongation factor 1a||CA-M||CTTGGAAAGGGATCGTTCAA||GCCTGTGAGGTTCCAGTGAT||||||||OK||Endoderm/Ectoderm diff. Scr. Larval Cdna
| |
| |-
| |
| |ENDO 16||Calcium Binding Protein||CL||GACCGAACGCCGATATAAGA||GCCATCGTCCCTTTAGTTCA||20/20||60.06/60.07||198bp||works well; clean disassociation curve||NCBI: L34680
| |
| |-
| |
| |Erg||Mesoderm ets factor||QT||AACGAGAGCCACATCTGGAG||TTTCGCTACGCTACAATCCA||||||150bp||||
| |
| |-
| |
| |EVE||Even Skipped||AR||CACAGACCCTGGACTTTCGT||GACAAACGGTCATCCCACTT||20/20||60.2/59.8||175 bp||works well; clean disassociation curve||cDNA clone from 12/15hr arrayed lib.
| |
| |-
| |
| |Ferr||Ferritin||CA-M||GCCTCGAGGAAGTCAGTCAT||ACCAACGTGGAGGTAGCATC||||||||OK||Endoderm/Ectoderm diff. Scr. Larval Cdna
| |
| |-
| |
| |FGF||Ligand||JN||CTCTTTGCCACCCTCATCAT||CCCTCGACTTGATGCTTTTC||20/20||~60/~60||172bp||works well; clean disassociation curve||
| |
| |-
| |
| |Ficolin||fibrinogen-domain protein||JR||GATGGACAAAGAGGGCTACG||TGAACCAGTCCGTCACTTCA||20/20||59.69/60.29||197||||CDNA
| |
| |-
| |
| |Fkh1/FoxB||SpFkh1||PO||AAGCCATCCACAACCAAATC||CATATCCGTCGAACGAGTCA||20/20||59.80/59.67||147bp||works well, clean dissociation curve||David,E.S. et Al (1999) NCBI:AF149706
| |
| |-
| |
| |FMO||Flavin-containing monooxygenase||CC||GTCGGTGGACAATCACCTCT||ACGAGGACATTCTTGCCATT||20/20||59.97/59.56||199||works well, clean dissociation curve||cDNA from 20 hrs arrayed lib.
| |
| |-
| |
| |FoxA||(=HNF3beta) TF||PO||CCAACCGACTCCGTATCATC||CGTAGCTGCTCATGCTGTGT||20/20||60.34/60.23||160bp||works well, clean dissociation curve||Coding from BAC clone
| |
| |-
| |
| |FoxN2/3||PMC TF||||TTCATGTCGATAGAGGACTGC||TTCGGAAGCACTTGTTGAGA||||||||||
| |
| |-
| |
| |FoxY||Forkead class gene FoxY||AR||TGCACTGCACTGACTCTGC||CTTTCCATTCCGTGGTGAAG||19/20||||||works well; clean disassociation curve||cDNA clone from 12 hr arrayed lib.
| |
| |-
| |
| |GAPDH||Phosphate DeHydrogenase internal standard||Garry Wessel||AGGCTTCTTCAGACGGACAG||TGCTAAGGCTGTTGGAAAGG||20/20||59.6/90.38||120||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |- | |
| |GATA c||TF||PO||CAGGGACATCATGTGCAAAC||CCGTGTTTGAATGCCTTCTT||20/20||59.97/60.11||155bp||works well, clean dissociation curve||cDNA clone from 20hr arrayed lib.
| |
| |-
| |
| |GATA e||Gata 4/5/6 factor||CL||ATGCATGCGGTCTCTACCA||CGCCACAGTGTTGTAGTGCT||19/20||||||works well; clean disassociation curve||NCBI: AF077675
| |
| |-
| |
| |GATA e (2)||TF||PO||CTGGCTCAAGACGAGAAGGA||CCTCTTCCGAGTCTGAATGC||20/20||60.67/59.95||176bp||works well, clean dissociation curve||cDNA clone from 20hr arrayed lib.
| |
| |-
| |
| |GCM||Glial Cells Missing||AR||CGACTGATAACCACGCTCAA||TTAACGACGTCGGTCGATTC||20/20||59.9/61.0||178 bp||works well; clean disassociation curve||cDNA clone from 12 hr arrayed lib.
| |
| |-
| |
| |Gelsolin||gelsolin-domain protein||JR||CTCCATCGACGAGAGGAGAA||CCTTCTGCTACGACCGAAAC||20/20||||||||
| |
| |-
| |
| |GFP||||PO||AGGGCTATGTGCAGGAGAGA||CTTGTGGCCGAGAATGTTTC||20/20||59.97/60.64||152bp||works well, clean dissociation curve||Cathy seq on EpGFP
| |
| |-
| |
| |Hedghog||Ligand||PO||ggcttcgattgggtcaacta||GTTGACCACGGCTACCTCAT||20/20||~60/~60||~150bp||works well; clean disassociation curve||
| |
| |-
| |
| |HES||Hairy||JR||TCTCAGGATTGGCAGCAAGT||CGTTAATCCTCGCTCGTCTT||20/20||60.94/59.48||175||||Cdna
| |
| |-
| |
| |HesC||Repressor of micromeres||||ccagaacagggcgaatctaa||CGAAGACGGGTTTCAATGTC||20/20||~60/~60||||||
| |
| |-
| |
| |Hex (TF 263)||PMC and SMC TF||MH||TTCTTGTGGAACCCGTTCAT||CGGGGAGAGGTATTTCTGGT||||||||||
| |
| |-
| |
| |Hmx||SpHmx||CL||TCGTCGTTTGAAGGTTGAAGT||TGATAGACGCATCTTGCTCG||21/20||59.77/60.12||152||works well; clean disassociation curve||NCBI: D85079
| |
| |-
| |
| |HNF3beta||HNF3 beta (same gene as FOXa)||AR||CATTGATCGTATCCGTGCTG||TTGCCACCGTTGTTGATTT||20/19||60.1/60.0||190 bp||works well; clean disassociation curve||cDNA clone from 15 hr arrayed lib.
| |
| |-
| |
| |HNF-6||Hepatocyte Nuclear Factor 6||OO||TGCAGCTTCTCTGCATACCA||ACTCCAACATGCCTCCAAAC||20/20||51.8/51.8||152bp||works well, clean dissociation curve||cDNA clone from 7/20/40hr arrayed lib.
| |
| |-
| |
| |Hox11/13B||TF Hox cluster||PO||CACAGGCTCTCGACCTAACC||GGTGGATGAGGTGGTAGATGA||20/21||59.87/59.79||155bp||works well, clean dissociation curve||Dobias et al(1998) NCBI:AF042652
| |
| |-
| |
| |Hsp70||Heat sock protein||CA-M||CACTTGGGTGGTGAGGACTT||TACCCTCAAACAGGGAATCG||||||||OK||Endoderm/Ectoderm diff. Scr. Larval Cdna
| |
| |-
| |
| |ISP1||nontrans poly A mRNA||AR||TGTGTTTCATTCCGTGGCTAT||GCCAACCCTTCTGATCAACT||21/19||60.4/59.1||180 bp||works well; clean disassociation curve||Calzone et al 1988; NCBI: Y00216
| |
| |-
| |
| |KAKAPO||actin binding||JR||GTGGCATTTATGAGCGGTCT||CGGCCCAGTACTTCAAGGAGA||20/20||60.10/59.28||177||OK||Bra screen
| |
| |-
| |
| |Krl||The original information has been deleted by someone, unknown reason.||TM (originally contributed by CBL)||CACGAACTCTTCGCAATCAA||CCAAGGGACAGGAGTGAAGA||||||||||
| |
| |-
| |
| |Lefty||Lefty, Nodal inhibitor||JN||CGTAGTCGCCACATCAGAGA||CAGATACATCATGGGCAACG||20/20||~60/~60||132bp||works well; clean disassociation curve||
| |
| |-
| |
| |LIM||Lim-HD||PO||GTATCCGATCCGTTGACGAC||TAGCCTTGCATTCACAGCAC||20/20||60.35/60.02||153bp||works well, clean dissociation curve||cDNA clone from 15hr arrayed lib.
| |
| |-
| |
| |Lys||Lysosomal associated protein||CA-M||TACCCGACAACCACTGTGTC||GCTCCCTCTCTGCGAAATAA||||||||OK||Endoderm/Ectoderm diff. Scr. Larval Cdna
| |
| |-
| |
| |Msp130||PMCspecific||GA||agagcaacgctcattctggt||ctccgaattgcattttgtca||20/20||60/59.7||149bp||seems ok: ask me for dissociation peak||
| |
| |-
| |
| |Nk1||Nkx1 homeobox TF||JR||GACCATGCATGTGCGTAAAC||TCTGTGACTGCCACTCATCC||20/20||60.00/59.83||175||works well; clean disassociation curve||Cdna
| |
| |-
| |
| |NKX 2.1||Apical plate from 24h on||GA||CGTGAGAGCTTCCCTACCTG||GAAGCTCCCTAGCTCGATGA||20/20||59.5/60.0||201bp||works well, clean dissociation curve||Peterson et al., umpublished data
| |
| |-
| |
| |Nodal||nodal, OE activator||JN||GACAACCCAAGCAACCACG||CGCACTCCTGTACGATCATG||19/20||~60/~60||178bp||works well with Sp, Lv, and Pl Nodal; clean disassociation curve||
| |
| |-
| |
| |Not||SpNot||CL||GAGCGACTTGAGCAGGAGTT||GGACCTGCTGTTTCTCGAAG||20/20||58.75/59.99||159||works well; clean disassociation curve||NCBI: AF109903 | |
| |-
| |
| |Notch||||CC||ACGGAGCCAAGCCTAAGAA||TCGTCACAGGCAACGAATAA||19/20||59.9/60.2||207||works well, clean dissociation curve||
| |
| |- | |
| |nrl||neuralized-like Zn fing; Ubq Ligase||JR||ATAGGTGCCCTGCACATAGG||ATCCAGCTTCCGAGGGTAAC||20/20||59.98/60.46||148||works well; clean disassociation curve||Cdna
| |
| |-
| |
| |OrCT||Endomeso diff gene||JR||GAC TTC AGA CGC GTT GGT CT||TTA TCA CTG TCG GGG AGT TTG||||||||||
| |
| |-
| |
| |Otx-alpha new||SpOtx-alpha (exon6)||CL/CY||CCTTACCAGCACCTGATCG||CTGGTCCTGCTGAACAAGGT||19/20||57.0/57.2||192||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Otx-beta1||This primer recognize both SpOtx-beta1and beta2 (exon 3 and 5)||CL/CY||GTTTAGGAACGTCGCTGGAA||TGAAGGTGGTGGTGATGTTG||20/20||55.15/55.15||487/187||The efficiency of this primer has been confirmed by CY. The previous Otx-beta primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Otx-beta2||Recognize exclusively SpOtx-beta1 (exon 4)||CL/CY||TGAATAACAGCCCTAGAAGAGCA||CTGCTCTACCGTCACCGATT||23/20||55.74/56.23||130||The efficiency of this primer has been confirmed by CY. The previous Otx-beta primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Otx-beta3||This primer recognize SpOtx-beta3 (exon 1 and 2)||CL/CY||CCACTCCACCGCTTCTACAC||CTTCAAGGTGCCGATAATTGA||20/21||59.25/53.42||150||The efficiency of this primer has been confirmed by CY. The previous Otx-beta primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Otx-beta4||This primer recognize SpOtx-beta1, beta2 and beta3 (exon 5)||CY||TGGATCATTCTGCCTTGACA||ACATAGCGGGATGCATGAG||20/19||54/54.9||189||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Pitx2||TF||MH||ACATTTCACCAGCCAGCAAC||TCAAGTTACACCACGCACAGA||||||104||||
| |
| |-
| |
| |PKS||polyketide synthase||JR||ATCGTTGGATCCTCAACAGC||GACACACTGTGCGCAATACC||20/20||60.08/60.19||195||works well; clean disassociation curve||Cdna
| |
| |-
| |
| |Pm27||PMCspecific||GA||cgaaagtggtctgctgatga||ctcgctctctctttccaacg||20/20||59.98/60.27||149bp||works well, clean dissociation curve||
| |
| |-
| |
| |Pmar1||Sp Hbox 12 Homeodomain TF||PO||GCGTTCAACGACAACCAGTA||GGTTGATGAGCAGAGCTTGA||20/20||59.76/59.12||152bp||works well, clean dissociation curve||cDNA clone from 9.5hr phage lib.
| |
| |-
| |
| |Prox1||SMC TF||||AGGTACCGGAGGGCTTCTT||ATGGTCTTCTTCCAGGATGG||||||||||
| |
| |-
| |
| |RFP||RFP (I got RFP clone from Mr. Damle)||JN||ATGAGGCTGAAGCTGAAGGA||TGGTGTAGTCCTCGTTGTGG||20/20||~60/~60||142bp||works well; clean disassociation curve||
| |
| |-
| |
| |S403||||CC||CTGCGGTGAGAGCAAGTTTA||GAGAGCGTTGCCTGAACAAC||20/20||59.2/60.99||199bp||works well, clean dissociation curve||cDNA from 20 hrs arrayed lib.
| |
| |-
| |
| |Six1/2||Aboral SMC TF||SM||TGAAACACCGTCAAAACAAGG||TGCTCTGGATGAAGATGAAGG|||||||||| | |
| |-
| |
| |SM30||Spicule Matrix protein||OO||GTTCTCCGGTAGGCAAACA||ACATTTTGGGGCAAATGAAA||19/20||59.4/59.9||196bp||works well, clean dissociation curve||Akasaka,K. ET AL.,1994.NCBI U05962
| |
| |-
| |
| |SM50-LC||SM50||LC||tagcctttgctacgggtcaa||ctgaggcgacgaaactgaa||20/19||60.4/60.16||||||
| |
| |-
| |
| |Snail||EMT TF expressed in SMC||SM||AAGGAGTACTCGACGTTCGG||TCCTGATGTGCATTTTGAGG||||||136bp||||
| |
| |-
| |
| |Sox B1||SpSox B1||CL||ATTCTGTGAACGTCATGGCA||TTGTCCTCTTGACCACACCA||20/20||60.12/60.13||184||bad dissossiation curve||NCBI: AF157389
| |
| |-
| |
| |Sox E||Sp SoxE||MH||CGGGAAGAGAAAACCTCACA||TTTTCCCAGGGTCTTGCTC||||||130bp||||
| |
| |-
| |
| |SoxC||Wave TF||MH||CATGGTTTGGTCACAAATCG||TACGGAGATTTCGCCACTTC||||||||||
| |
| |-
| |
| |Sp cbf-a||CCAAT-binding factor, A (Ubq expression)||TB||ACCCATCGCTAATGTTGCTC||CACTGGCTTCGCTTGTGATA||20/20||60/60||131||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp cbf-b||CCAAT-binding factor, B (Ubq expresion)||TB||GGATTCCCAAGGAGAGAAGG||ATAGAGGGCGAAGGGATCAT||20/20||60/60||130||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp cbf-c||CCAAT-binding factor, C (Ubq expression)||TB||TCCTCTTATTCTCTTCTGTATGTAGCC||ATAAGTGCTGAAGCCCCAGT||27/20||60/60||100||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp gcf-1||GCF1, general activator||TB||ACCAGACCTTCTCCCACCTT||TCTCTTGCGGTTGTTCTCCT||20/20||60/60||||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp myb||myb (cyIIIa repressor)||TB||AACCATTCACAAGCCACTCC||TGGTCCTCCTGCCTTCTCTA||20/20||60/60||147||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp oct-1||oct-1, cyIIIa activator||TB||CACAGGGTGATGTTGGTCTG||GGAGAGGTTTGAGCTTGCAC||20/20||60/60||123||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp P3A2||P3A2, OE repressor of cyIIIa||TB||AGCATCATGGAAGGGATGAC||GTGTACCACAGCATGGGATG||20/20||60/60||104||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp runt||runt (cyIIIa activator, ubiq)||TB||CGGTACGGAGGAACAACCTA||AGGGCTCTCTGTCTTCACCA||20/20||60/60||149||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp tef-1||TEF-1/scalloped, cyIIIa activator||TB||GGCCACTGCCTTACAAGAAC||AGGGTCATGAGATGGTCCTG||20/20||60/60||146||The efficiency of this primer has been confirmed by CY. The previous Otx-alpha primer has been removed from this data base because it does not work well.||
| |
| |-
| |
| |Sp tondu||tondu/vestigial, TEF-1 co-factor||TB||ACTCCGGTGATTTGGACAAG||CAGTGAGCGGCTAAAATGTTC||20.21||60/60||50||The efficiency of this primer has been confirmed by CY.||
| |
| |-
| |
| |The previous Otx-alpha primer has been removed from this data base because it does not work well.||||||||||||||||||
| |
| |-
| |
| |SpDlx||Aboral Ectoderm TF||EL||CCAGCTTACAACTCCAACAGC||TTACCTGAGTTTGAGTGAGTCCA||||||||||
| |
| |-
| |
| |SPEC 1||aboral ectoderm||GA||GCGCATGCTCAGATTGTATC||CAAGGAGTGGTAAGGGTGGA||20/20||58.6/59.0||164 bp||works well, clean dissociation curve||Hardin,S.H.,1985 NCBI X03287
| |
| |-
| |
| |Spec2a||SpSpec2a||PYL||GGACGGTAAAATCTGCCTTG||AGACGATTATGTCACCCTCTCC||||||||The efficiency of this primer has been confirmed by PYL.||
| |
| |-
| |
| |SpElk||Ets TF||Inna||ATCATGGTCGCTAGTCCTCTCCT||TGACAAGAGAACAGTCGGTGTGA||||||||||
| |
| |-
| |
| |SpEts||TF||PO||GCACTGGTCCATCAAGGAGT||GATGTGCTCCCAGAGGATGT||20/20||60.12/60.08||145bb||works well, clean dissociation curve||Rao and Childs(1993) NCBI:L19541
| |
| |-
| |
| |SpEts4||TF Ets4||PO||CTCCAGCCCAACTCCTACAG||GATGGAGCGAGAGAGCTTGT||20/20||60||||works well, checked by Paola||
| |
| |-
| |
| |SpGsc||Homeodomain TF||PO||GCGACACGCTCCCTATCTAC||CGATGTCGCCTCTTTCTCTT||20/20||59.87-59.57||147bp||works well, clean dissociation curve||Angerer(2001)-NCBI: AF315231
| |
| |-
| |
| |SpIrxA||Aboral Ectoderm TF||EL||TATGGAATGGACCTGAACGG||TATGATCTTTTCGCCCTTGG||||||||||
| |
| |-
| |
| |SpLox||A gene that is expresed in the endoderm at 45 h||Ina Arnone||GTGCGACGGACTCCCTATAA||TTCAGACGCCATGGTGTAAA||20/20||56.2/54.4||||works well, clean dissociation curve||
| |
| |-
| |
| |SpMSP130like||PMC differentiation gene||JR||TTC TTG GTC GCC TGG ATT AC||ACC TTG GCA TCG CAT AGA AG||||||||||
| |
| |-
| |
| |SpNK2.2||Aboral Ectoderm TF||EL||ACACTTGGCGAGCATTATCC||CGGAGAAGGTAACGGATTCA||||||||||
| |
| |-
| |
| |SpTbx2/3||Aboral Ectoderm TF||EL||ACTGCCGGTACAAGTTCCAC||GACACATTTCTGCATCCATTG||||||||||
| |
| |-
| |
| |SpZ12-1||TF||PO||AGTCGTCCAGCCATGTCTTT||AAGCACACCTCGCACCTATC||20/20||59.73/60.29||151bp||works well, clean dissociation curve||Wang D. et Al (1995) NCBI:U19831
| |
| |-
| |
| |Su(H)||Suppressor of hairless||CC||GCTCCATCGTTGATGATCTCT||GGACGCTGATGATCCAGTCT||21/20||59.26/60.23||156||works well, clean dissociation curve||partial seq from Posakony's lab
| |
| |-
| |
| |SuTx||sulfotransferase||CC||AATTCATGCCAGAGCCATTG||CCGAGAACTCGACCTTCAAC||20/20||61/59.84||204||works well, clean dissociation curve||cDNA from 20 hrs arrayed lib.
| |
| |-
| |
| |TBR||T-brain||JR||GAAACATTCGCCTTCCTTGT||GAAGGCGTCGGTTTACCTCT||20/20||59.17/60.63||98||primers reversed, and 3' primer bridges exon1 and exon2, making this pair unsuitable for genomic DNA quantitation||Cdna
| |
| |-
| |
| |Tel||Maternal + Zygotic mesoderm TF||QT||TGTTAGCTTCTGCCCCTGTT||CGACGAGAGAGGGTCTTCAG||||||162 bp||||
| |
| |-
| |
| |Tgif (ark 43)||PMC and Endoderm TF||MH||GCTCTACCTATCTCGCTTGGC||TGGTGAACTTGTCAGGGTCT||||||||||
| |
| |-
| |
| |UBQ||Ubiquitin||CL||CACAGGCAAGACCATCACAC||GAGAGAGTGCGACCATCCTC||20/20||60.16/59.95||147bp||works well; clean disassociation curve||
| |
| |-
| |
| |VegFR||VegF Receptor||JR||AGGGCAGGTCAACAGTTCAG||GGCAACCAATTTGACATCCT||||||||||
| |
| |-
| |
| |Wnt 5a||Wnt 5a||PYL||TGCTGTGGAAGAGGCTACAA||TTCTGCACTTCCGACACTTG||20/20||||||works well, clean dissociation curve||
| |
| |-
| |
| |WNT 8 (2)||||PO||TGTCGTTCATTCAAGCCATC||TATCACTCGCCATTCGTTCA||20/20||59.65/60.22||183bp||works well, clean dissociation curve||
| |
| |} | | |} |