Bioinformatic Resources and BAC resources: Difference between pages
imported>Ctelmer m (Ctelmer moved page Bioinformatic resources to Bioinformatic Resources: There was not a link to the Main Page.) |
imported>Echinobase |
||
| Line 1: | Line 1: | ||
== Genomic BAC Resources == | |||
Echinobase stores libraries of genomic sequence fragments maintained in bacterial artificial chromosome (BAC) vectors for seven lower deuterostome species [[Echinobase/bac_table/bac_table.php|'''(BAC Table)''']]. These libraries were originally constructed as part of the Sea Urchin Genome Project. On average, the insert sizes for our BAC libraries are ~140 kb. Thus, for the 800 Mb ''S. purpuratus'' genome, 1X coverage is encompassed by, on average, 5,700 clones. Each library consists of >100,000 clones, providing ~17X genome coverage. | |||
A subset of the ''S. purpuratus'' BACs have been mapped to the genome assembly. As part of the SUGP, about 8,000 BAC clones were end sequenced using Sanger technology. Genomic positions for these BACs are indicated as a track in JBrowse (link). An additional set of individual BACs were sequenced completely; links to the NCBI page is provided for these. Finally, some BACs have been identified using hybridization screening of arrayed library filters (see [[Echinobase/bac_table/bac_table.php|'''(BAC Table)''']]) | |||
[http:// | '''BAC Vector Sequence'''. The BACs are maintained in the [http://bacpac.chori.org/pbace36.htm pBACe3.6 vector] which originates from Children's Hospital Oakland Research Institute in Oakland, California, USA. This vector confers resistance to the antibiotic chloramphenicol and is fully described by [http://www.ncbi.nlm.nih.gov/pubmed/10373322?dopt=Abstract Frengen and colleages]. The vector sequence is available [http://www.ncbi.nlm.nih.gov/nuccore/4878025?report=fasta here]. | ||
[http:// | '''Relevant publications:'''<br /> | ||
[http://www.pnas.org/content/97/17/9514.full A sea urchin genome project: Sequence scan, virtual map, and additional resources.] Cameron, RA, et al., 2000. Proc. Natl. Acad. Sci. USA.<br /> | |||
[[LINK. Bacterial artificial chromosomes as recombinant reporter constructs to investigate gene expression and regulation in echinoderms.]] Buckley, KM et al., 2017. Briefings in Functional Genomics. | |||
== BAC Protocols == | |||
Screening a BAC library<br /> | |||
Generating a recombinant BAC<br /> | |||
BAC miniprep protocol<br /> | |||
Analyzing BAC DNA by PFGE | |||
<br /> | |||
== Recombinant BAC Resources == | |||
Echinobase has generated and maintains a set of reporter BAC constructs in which a portion of the gene of interest is replaced by the sequence encoding a fluorescent protein (''i.e.'' GFP, mCherry). A list of these sequences is available [[Echinobase/bac_table/bac_table.php|'''here''']]). Existing clones are available by request. We will also consider requests to make additional recombinant reporter constructs as time permits. | |||
'''To request a recombinant or wildtype BAC for non-commercial use:'''<br /> | |||
1. Identify the BAC of interest computationally or by screening our libraries ([[Echinobase/filters|available filters]]).<br /> | |||
2. Request the BAC by emailing [mailto:kbuckle2@andrew.cmu.edu Kate Buckley] or [mailto:veronica@cmu.edu Veronica Hinman]. | |||
<br /> | |||
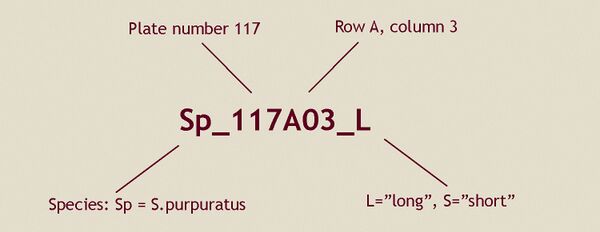
[[File:Bac.jpg|600px]] | |||
== Working with BACs == | |||
'''Storing BACs''' | |||
'''Short term'''<br /> | |||
To avoid shearing the long BAC DNA fragments, ''isolated BACs should never be vortexed or frozen''. Store BAC DNA at 4 C in TE (10 mM Tris, pH = 7.4; 10 mM EDTA).<br /> | |||
'''Long term storage'''<br /> | |||
Store glycerol bacteria stocks (15-25% glycerol) at -80 C. | |||
'''Antibiotic conditions for working with BAC clones'''<br /> | |||
'''Kanamycin'''<br /> | |||
Stock (25 μg/μL): Dissolve 0.5 g of kanamycin into 10 ml of ddH2O. Filter through a 0.22 µm filter to sterilize. Aliquot and store at -20°C.<br /> | |||
Use at 1:1000 dilution (25 μg/mL).<br /> | |||
'''Chloramphenicol'''<br /> | |||
Stock (25 μg/μl): Dissolve in ETOH store at -20.<br /> | |||
Use at 1:2000 dilution.(12.5 μg/ml) | |||
Revision as of 18:31, 12 February 2020
Genomic BAC Resources
Echinobase stores libraries of genomic sequence fragments maintained in bacterial artificial chromosome (BAC) vectors for seven lower deuterostome species (BAC Table). These libraries were originally constructed as part of the Sea Urchin Genome Project. On average, the insert sizes for our BAC libraries are ~140 kb. Thus, for the 800 Mb S. purpuratus genome, 1X coverage is encompassed by, on average, 5,700 clones. Each library consists of >100,000 clones, providing ~17X genome coverage.
A subset of the S. purpuratus BACs have been mapped to the genome assembly. As part of the SUGP, about 8,000 BAC clones were end sequenced using Sanger technology. Genomic positions for these BACs are indicated as a track in JBrowse (link). An additional set of individual BACs were sequenced completely; links to the NCBI page is provided for these. Finally, some BACs have been identified using hybridization screening of arrayed library filters (see (BAC Table))
BAC Vector Sequence. The BACs are maintained in the pBACe3.6 vector which originates from Children's Hospital Oakland Research Institute in Oakland, California, USA. This vector confers resistance to the antibiotic chloramphenicol and is fully described by Frengen and colleages. The vector sequence is available here.
Relevant publications:
A sea urchin genome project: Sequence scan, virtual map, and additional resources. Cameron, RA, et al., 2000. Proc. Natl. Acad. Sci. USA.
LINK. Bacterial artificial chromosomes as recombinant reporter constructs to investigate gene expression and regulation in echinoderms. Buckley, KM et al., 2017. Briefings in Functional Genomics.
BAC Protocols
Screening a BAC library
Generating a recombinant BAC
BAC miniprep protocol
Analyzing BAC DNA by PFGE
Recombinant BAC Resources
Echinobase has generated and maintains a set of reporter BAC constructs in which a portion of the gene of interest is replaced by the sequence encoding a fluorescent protein (i.e. GFP, mCherry). A list of these sequences is available here). Existing clones are available by request. We will also consider requests to make additional recombinant reporter constructs as time permits.
To request a recombinant or wildtype BAC for non-commercial use:
1. Identify the BAC of interest computationally or by screening our libraries (available filters).
2. Request the BAC by emailing Kate Buckley or Veronica Hinman.
Working with BACs
Storing BACs
Short term
To avoid shearing the long BAC DNA fragments, isolated BACs should never be vortexed or frozen. Store BAC DNA at 4 C in TE (10 mM Tris, pH = 7.4; 10 mM EDTA).
Long term storage
Store glycerol bacteria stocks (15-25% glycerol) at -80 C.
Antibiotic conditions for working with BAC clones
Kanamycin
Stock (25 μg/μL): Dissolve 0.5 g of kanamycin into 10 ml of ddH2O. Filter through a 0.22 µm filter to sterilize. Aliquot and store at -20°C.
Use at 1:1000 dilution (25 μg/mL).
Chloramphenicol
Stock (25 μg/μl): Dissolve in ETOH store at -20.
Use at 1:2000 dilution.(12.5 μg/ml)