QPCR and Array: Difference between pages
imported>Ctelmer (→QPCR) |
imported>Ctelmer |
||
| Line 1: | Line 1: | ||
<div class="content"> | |||
<div class="field field-name-body field-type-text-with-summary field-label-hidden"> | |||
<div class="field-items"> | |||
<div class="field-item even" property="content:encoded"> | |||
These procedures are our standard operating procedure. They are derived from the original procedures published on the net by Hans Lehrach's lab in Berlin and include our modifications. | |||
'''Table of Contents''' | |||
# [[#screen|How To Screen Filters]] | |||
# [[#microwell|How To Determine Microwell Plate Coordinates From The Arrayed Filter]] | |||
# [[#filters|How To Strip Filters]] | |||
=== HOW TO SCREEN FILTERS === | |||
1. Make up a pre-hybridization solution as follows (for 25 ml) | |||
6.25 mlof 20x SSPE (to a conc. of 5x SSPE) | |||
0.5 ml of 5% PPi (final conc. 0.1%) | |||
1.25 ml of 100x Denhardt's solution (to a conc. of 5x Denhardt's solution) | |||
0.625 ml of 10% (w/v) SDS (to a conc. of 0.5% (w/v) SDS) | |||
16.37 ml of dH<sub>2</sub>O | |||
Make up to 24.5 ml with sterile water. Add to the membrane in a hybridization box or bag. In the past, we have used a lucite box that is 11X11x6 inches in a New Brunswick shaking water bath. Recently,we have moved to Hybaid hybridization ovens with glass bottles for hybridization. A complete set of library filters will fit in one bottle for both hybridization and washing. Follow the manufacturer's instructions for handling bottles. | |||
2. Denature 0.5 ml of a 1 mg/ml solution of sonicated blocking DNA by heating to 100°C for 5 min. Chill on ice and add to prehybridization solution. | |||
3. Prehybridize in a shaking water bath at 65°C for 1 hr. | |||
4. Denature labeled probe (unless using an RNA or ssDNA probe) by heating to 100°C for 5 min. Add the probe to the prehybridization solution. | |||
5. Incubate for at least 12 hr at 65°C. | |||
6. Following hybridization, wash the filters by incubating them in 2x SSPE, 0.1% (w/v) SDS at room temperature for 10 min. Repeat. | |||
7. Replace the solution with 1x SSPE, 0.1% (w/v) SDS. Incubate at 65°C for 15 min. | |||
8. Replace the solution with 1x SSPE, 0.1% (w/v) SDS. Incubate at 65°C for 10 min. Repeat if necessary. Note: This is a moderately high stringency wash and should be omitted if related sequences are to be probed. The stringency can be increased to 0.1X SSPE, 0.1% SDS for high stringency conditions. | |||
| | |||
9. Remove filter, wrap in plastic wrap and carry out autoradiography. We routinely use <sup>32</sup>P probes which pass through plastic wrap with little loss. Plastic wrap results in loss of much of the signal when <sup>35</sup>S probes are used. Without plastic wrap contamination is a severe problem so we've avoided using <sup>35</sup>S probes. Plastic wrap should be employed without trapped air for best exposures. From the final wash, pick up the filter by one corner and allow it to drip dry for 10 seconds. Place the filter face up on a piece of plastic wrap still attached to the roll. Fold the attached edge off the filter, then roll the top layer of wrap onto the filter. This expels any trapped stripped. | |||
[[#top|Back to the top of the page]] | |||
=== HOW TO DETERMINE MICROWELL PLATE COORDINATES FROM THE ARRAYED FILTER === | |||
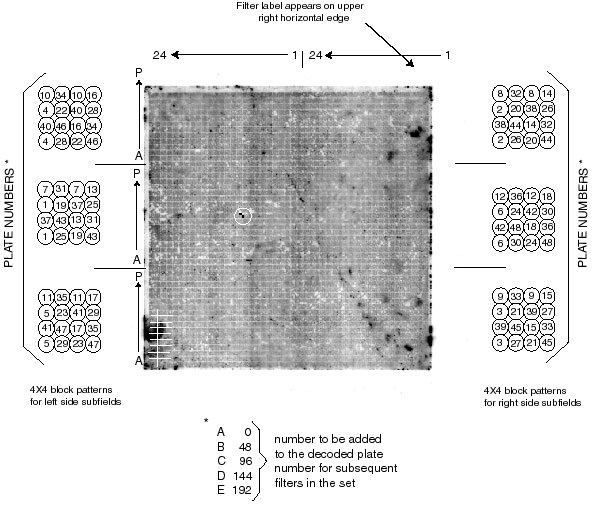
The high-density filter array is a square arrangement of 48X48 blocks which can be thought of as six sub-fields of 16X24 blocks. Thus each sub-field is equivalent to the wells of a 384-well plate. Each block is a 4X4 array of eight clones spotted in duplicate. That is, the inoculum from each well of each plate has been spotted twice onto the filter in the same 4X4 block. The arrangement has been designed so that the two spots define a unique angle different from all the others within the 4X4 block (see examples in the figure below). The unique angular relationship of the spot pair defines the plate from which that clone was taken. In the accompanying figure, a 4X4 block adjacent to each sub-field indicates the plate number assignments for the blocks in that sub-field. The position of a 4X4 block containing a positive spot pair can be described by the X-Y coordinates of the block in the sub-field (X coordinates are A through P, from bottom to top; Y coordinates are 1 through 24, from right to left). For filters beyond the first one (A) in the set, the plate numbers are increased in increments of 48, thus the plate number for the B filter is the decoded number plus 48; for the C filter, plus 96; etc. | |||
EXAMPLE: On the accompanying figure there is a positive spot pair circled in white. It lies in the left-middle sub-field at X-Y position I-10. That is I blocks (9) up from the bottom of the sub-field and 10 blocks over from the centerline. Thus the well position with in the plate is I-10. For this sub-field, the angle of the spot pair within the block indicates #31. There fore, the clone is located on plate #31 in well I-10. | |||
NOTES: Identification of spot coordinates in the case where the background on the filter is very low is aided by pre-marking the filters when they are dry and the colony residue is visible. Dry filters are marked by indentations from a ball point pen. The pen tip is pressed into a filter that is placed on a piece of 3MM paper on a hard surface while observing the operation in oblique lighting. Dots can thus be made at the boundaries of the 6 sub-fields and at the extreme corners of the array. | |||
To aid orientation in the newer filter sets, the A1 well for each plate has been left empty. Thus there are six empty squares that have no bacteria on the filter. After hybridization these squares have lower background and will orient the filter. The squares are 3 sets of 2 across when the label is on the upper right hand edge. | |||
<p style="margin-left: 20px">[[File:Filter_coordinates.jpg|frameless|1000px]]</p> | |||
=== THE Sp FILTER COLLECTION === | |||
<table> | |||
<th> R number | |||
<th> Library name | |||
<th> Notes | |||
<tr> | |||
<td>1-192 | |||
<td> BAC Sp 8/98 | |||
<td> original library construction made in 1998 | |||
<tr> | |||
<td>1000-1152 | |||
<td> BAC Sp FR2 | |||
<td> 1-40 and 97-152; actually 1000 so 1006 means Fr2 #6</tr> | |||
<tr> | |||
<td>3000-3123 | |||
<td> BAC Sp Cond | |||
<td> Cond = condensed. A set of condensed filters derived from 8/98 and FR2</tr> | |||
<tr> | |||
<td>4000-4144 | |||
<td> BAC Sp Ext | |||
<td> A second construction to find rare clones</tr> | |||
<tr> | |||
<td>5000-5330 | |||
<td> BAC Sp Mbo | |||
<td> Another library made with Mbo instead of EcoRI</tr> | |||
</table> | |||
[[#top|Back to the top of the page]] | |||
=== HOW TO STRIP FILTERS === | |||
; 1.Incubate the blot at 45°C for 30 min in 0.4 M NaOH (80 ml of 5 M NaOH, 920 ml of H<sub>2</sub>O).<br /> | |||
2.Transfer the membrane to the stripping buffer: | |||
: 0.1x SSC (5 ml of 20x SSC) | |||
: 0.1% SDS (w/v) (5 ml of 20% SDS) | |||
: 0.2 M Tris-HCl, pH 7.5 (200 ml of 1 M Tris, 790 ml H<sub>2</sub>O) | |||
: Incubate at 60°C (can be increased up to 65°C) for 15-30min. | |||
; 3.Transfer the membrane to 20 mM EDTA containing stripping buffer: | |||
: 96 ml of the stripping buffer | |||
: 4 ml of 0.5 M EDTA | |||
: Incubate at temperature for at least 10 min. | |||
; 1) Short-term storage (1 or 2 weeks) | |||
: a. Wet two sheets of Whatmann paper in EDTA-containing stripping buffer. | |||
: b. Place the membrane between the two wet papers. | |||
: c. Wrap the papers and the membrane with plastic wrap and keep them in refrigerator until reuse. | |||
; 2) Long-term storage | |||
: a. Sandwich between two sheets of plastic wrap. | |||
: b. Expose the membrane to X-ray film for at least 12 hr to check whether stripping is done completely. If stripping is done completely, place the membrane between two sheets of dry Whatmann paper and dry it at room temperature for at least 24 hr(until completely dry). | |||
: c. If stripping is not complete, repeat steps 1&2 but execute step 2 at the higher temperature. | |||
; Alternative stripping method: | |||
: Bring 0.5% SDS to a boil. | |||
: Pour on the membrane and allow to cool to room temperature. | |||
: (If necessary, repeat) | |||
</div> | |||
</div> | |||
</div> | |||
</div> | |||
Revision as of 11:12, 9 June 2021
These procedures are our standard operating procedure. They are derived from the original procedures published on the net by Hans Lehrach's lab in Berlin and include our modifications.
Table of Contents
- How To Screen Filters
- How To Determine Microwell Plate Coordinates From The Arrayed Filter
- How To Strip Filters
HOW TO SCREEN FILTERS
1. Make up a pre-hybridization solution as follows (for 25 ml) 6.25 mlof 20x SSPE (to a conc. of 5x SSPE) 0.5 ml of 5% PPi (final conc. 0.1%) 1.25 ml of 100x Denhardt's solution (to a conc. of 5x Denhardt's solution) 0.625 ml of 10% (w/v) SDS (to a conc. of 0.5% (w/v) SDS) 16.37 ml of dH2O Make up to 24.5 ml with sterile water. Add to the membrane in a hybridization box or bag. In the past, we have used a lucite box that is 11X11x6 inches in a New Brunswick shaking water bath. Recently,we have moved to Hybaid hybridization ovens with glass bottles for hybridization. A complete set of library filters will fit in one bottle for both hybridization and washing. Follow the manufacturer's instructions for handling bottles.
2. Denature 0.5 ml of a 1 mg/ml solution of sonicated blocking DNA by heating to 100°C for 5 min. Chill on ice and add to prehybridization solution.
3. Prehybridize in a shaking water bath at 65°C for 1 hr.
4. Denature labeled probe (unless using an RNA or ssDNA probe) by heating to 100°C for 5 min. Add the probe to the prehybridization solution.
5. Incubate for at least 12 hr at 65°C.
6. Following hybridization, wash the filters by incubating them in 2x SSPE, 0.1% (w/v) SDS at room temperature for 10 min. Repeat.
7. Replace the solution with 1x SSPE, 0.1% (w/v) SDS. Incubate at 65°C for 15 min.
8. Replace the solution with 1x SSPE, 0.1% (w/v) SDS. Incubate at 65°C for 10 min. Repeat if necessary. Note: This is a moderately high stringency wash and should be omitted if related sequences are to be probed. The stringency can be increased to 0.1X SSPE, 0.1% SDS for high stringency conditions.
9. Remove filter, wrap in plastic wrap and carry out autoradiography. We routinely use 32P probes which pass through plastic wrap with little loss. Plastic wrap results in loss of much of the signal when 35S probes are used. Without plastic wrap contamination is a severe problem so we've avoided using 35S probes. Plastic wrap should be employed without trapped air for best exposures. From the final wash, pick up the filter by one corner and allow it to drip dry for 10 seconds. Place the filter face up on a piece of plastic wrap still attached to the roll. Fold the attached edge off the filter, then roll the top layer of wrap onto the filter. This expels any trapped stripped.
HOW TO DETERMINE MICROWELL PLATE COORDINATES FROM THE ARRAYED FILTER
The high-density filter array is a square arrangement of 48X48 blocks which can be thought of as six sub-fields of 16X24 blocks. Thus each sub-field is equivalent to the wells of a 384-well plate. Each block is a 4X4 array of eight clones spotted in duplicate. That is, the inoculum from each well of each plate has been spotted twice onto the filter in the same 4X4 block. The arrangement has been designed so that the two spots define a unique angle different from all the others within the 4X4 block (see examples in the figure below). The unique angular relationship of the spot pair defines the plate from which that clone was taken. In the accompanying figure, a 4X4 block adjacent to each sub-field indicates the plate number assignments for the blocks in that sub-field. The position of a 4X4 block containing a positive spot pair can be described by the X-Y coordinates of the block in the sub-field (X coordinates are A through P, from bottom to top; Y coordinates are 1 through 24, from right to left). For filters beyond the first one (A) in the set, the plate numbers are increased in increments of 48, thus the plate number for the B filter is the decoded number plus 48; for the C filter, plus 96; etc.
EXAMPLE: On the accompanying figure there is a positive spot pair circled in white. It lies in the left-middle sub-field at X-Y position I-10. That is I blocks (9) up from the bottom of the sub-field and 10 blocks over from the centerline. Thus the well position with in the plate is I-10. For this sub-field, the angle of the spot pair within the block indicates #31. There fore, the clone is located on plate #31 in well I-10.
NOTES: Identification of spot coordinates in the case where the background on the filter is very low is aided by pre-marking the filters when they are dry and the colony residue is visible. Dry filters are marked by indentations from a ball point pen. The pen tip is pressed into a filter that is placed on a piece of 3MM paper on a hard surface while observing the operation in oblique lighting. Dots can thus be made at the boundaries of the 6 sub-fields and at the extreme corners of the array.
To aid orientation in the newer filter sets, the A1 well for each plate has been left empty. Thus there are six empty squares that have no bacteria on the filter. After hybridization these squares have lower background and will orient the filter. The squares are 3 sets of 2 across when the label is on the upper right hand edge.
THE Sp FILTER COLLECTION
| R number | Library name | Notes |
|---|---|---|
| 1-192 | BAC Sp 8/98 | original library construction made in 1998 |
| 1000-1152 | BAC Sp FR2 | 1-40 and 97-152; actually 1000 so 1006 means Fr2 #6 |
| 3000-3123 | BAC Sp Cond | Cond = condensed. A set of condensed filters derived from 8/98 and FR2 |
| 4000-4144 | BAC Sp Ext | A second construction to find rare clones |
| 5000-5330 | BAC Sp Mbo | Another library made with Mbo instead of EcoRI |
HOW TO STRIP FILTERS
- 1.Incubate the blot at 45°C for 30 min in 0.4 M NaOH (80 ml of 5 M NaOH, 920 ml of H2O).
2.Transfer the membrane to the stripping buffer:
- 0.1x SSC (5 ml of 20x SSC)
- 0.1% SDS (w/v) (5 ml of 20% SDS)
- 0.2 M Tris-HCl, pH 7.5 (200 ml of 1 M Tris, 790 ml H2O)
- Incubate at 60°C (can be increased up to 65°C) for 15-30min.
- 3.Transfer the membrane to 20 mM EDTA containing stripping buffer
- 96 ml of the stripping buffer
- 4 ml of 0.5 M EDTA
- Incubate at temperature for at least 10 min.
- 1) Short-term storage (1 or 2 weeks)
- a. Wet two sheets of Whatmann paper in EDTA-containing stripping buffer.
- b. Place the membrane between the two wet papers.
- c. Wrap the papers and the membrane with plastic wrap and keep them in refrigerator until reuse.
- 2) Long-term storage
- a. Sandwich between two sheets of plastic wrap.
- b. Expose the membrane to X-ray film for at least 12 hr to check whether stripping is done completely. If stripping is done completely, place the membrane between two sheets of dry Whatmann paper and dry it at room temperature for at least 24 hr(until completely dry).
- c. If stripping is not complete, repeat steps 1&2 but execute step 2 at the higher temperature.
- Alternative stripping method
- Bring 0.5% SDS to a boil.
- Pour on the membrane and allow to cool to room temperature.
- (If necessary, repeat)