ECB-FEAT-23044680: Difference between revisions
Jump to navigation
Jump to search
Created page with "===<i>Strongylocentrotus purpuratus</i> Genome Assembly v5.0=== The gene model, LOC580257, appears to be incomplete, with the upstream locus, LOC115922204, most likely being..." |
|||
| Line 1: | Line 1: | ||
===<i>Strongylocentrotus purpuratus</i> Genome Assembly v5.0=== | ===<i>Strongylocentrotus purpuratus</i> Genome Assembly v5.0=== | ||
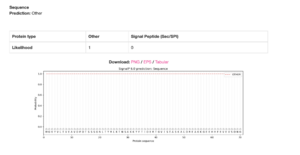
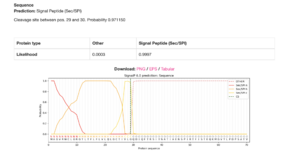
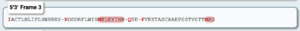
The gene model, LOC580257, appears to be incomplete, with the upstream locus, LOC115922204, most likely being part of this gene based on several lines of evidence. The lines of evidence for a split-gene misannotation include the lack of signal peptide sequences yielded by SMART (Letunic et al., 2006) and SignalP 6.0 analyses (Teufel et al., 2022), the absence of short open reading frames and stop codons within the 5’-UTR in the mRNA transcript, XM_030980581.1, yielded by the ExPASy Translate tool (Gasteiger et al., 2003), and the adjacency of the coding loci on scaffold NW_022145596.1. | The gene model, LOC580257, appears to be incomplete, with the upstream locus, LOC115922204, most likely being part of this gene based on several lines of evidence. The lines of evidence for a split-gene misannotation include the lack of signal peptide sequences yielded by SMART ([https://academic.oup.com/nar/article/49/D1/D458/5940513?login=true Letunic et al., 2006]) and SignalP 6.0 analyses ([https://www.nature.com/articles/s41587-021-01156-3 Teufel et al., 2022]), the absence of short open reading frames and stop codons within the 5’-UTR in the mRNA transcript, XM_030980581.1, yielded by the ExPASy Translate tool ([https://academic.oup.com/nar/article/31/13/3784/2904185 Gasteiger et al., 2003]), and the adjacency of the coding loci on scaffold NW_022145596.1. | ||
[[File:SMART LOC580257.png|thumb|Incomplete Model (LOC580257) for ''Strongylocentrotus purpuratus'' in SMART]] | [[File:SMART LOC580257.png|thumb|Incomplete Model (LOC580257) for ''Strongylocentrotus purpuratus'' in SMART]] | ||
Latest revision as of 19:13, 1 May 2023
Strongylocentrotus purpuratus Genome Assembly v5.0
The gene model, LOC580257, appears to be incomplete, with the upstream locus, LOC115922204, most likely being part of this gene based on several lines of evidence. The lines of evidence for a split-gene misannotation include the lack of signal peptide sequences yielded by SMART (Letunic et al., 2006) and SignalP 6.0 analyses (Teufel et al., 2022), the absence of short open reading frames and stop codons within the 5’-UTR in the mRNA transcript, XM_030980581.1, yielded by the ExPASy Translate tool (Gasteiger et al., 2003), and the adjacency of the coding loci on scaffold NW_022145596.1.