Genome Assemblies: Difference between revisions
No edit summary |
|||
| Line 27: | Line 27: | ||
The genome assembly is available on NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA63057 PRJNA63057]. | The genome assembly is available on NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA63057 PRJNA63057]. | ||
=='''''Mesocentrotus franciscanus'''''== | |||
Red sea urchins, Mesocentrotus franciscanus (A. Agassiz, 1863) range from Alaska to Baja California at depths from just below the low tide line to 90 m. It prefers rocky shores that are sheltered from strong wave action. Although the red urchin has long been called ''Strongylocentrotus franciscanu''s, recent molecular phylogenies support the name to ''Mesocentrotus franciscanus'' (A. Agassiz, 1863) (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88). | Red sea urchins, Mesocentrotus franciscanus (A. Agassiz, 1863) range from Alaska to Baja California at depths from just below the low tide line to 90 m. It prefers rocky shores that are sheltered from strong wave action. Although the red urchin has long been called ''Strongylocentrotus franciscanu''s, recent molecular phylogenies support the name to ''Mesocentrotus franciscanus'' (A. Agassiz, 1863) (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88). | ||
A low coverage sequence of this species is available at NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA20313 PRJNA20313]. | A low coverage sequence of this species is available at NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA20313 PRJNA20313]. | ||
=='''''Allocentrotus fragilis'''''== | |||
''Allocentrotus fragilis'', the fragile sea urchin, lives on the sea floor at depths from 300 to 1600 ft. While ''Allocentrotus fragilis'' has long been the accepted name used for this species in the family Strongylocentrotidae, a more appropriate name might be ''Strongylocentrotus fragilis'' (Jackson, 1912). Indeed, the molecular evidence fully supports this change of name (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88). | ''Allocentrotus fragilis'', the fragile sea urchin, lives on the sea floor at depths from 300 to 1600 ft. While ''Allocentrotus fragilis'' has long been the accepted name used for this species in the family Strongylocentrotidae, a more appropriate name might be ''Strongylocentrotus fragilis'' (Jackson, 1912). Indeed, the molecular evidence fully supports this change of name (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88). | ||
A low coverage sequence of this species is available at NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA20317 PRJNA20317]. | A low coverage sequence of this species is available at NCBI at accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA20317 PRJNA20317]. | ||
=='''''Amphiura filiformis'''''== | |||
''Amphiura filiformis'' (O. F. Muller, 1776) is a brittle star found on the seabed of the European coasts from Greece to Norway and Sweden including the Baltic Sea . It is an important member of soft bottom communities where it digs a shallow burrow and feeds on plankton. It serves as food for crayfish and finfish. Stemming from its remarkable ability to regenerate arms in a matter of weeks it has become an emerging model for regeneration and stem cell biology in biomedical research. Research targets include: 1) The cellular and molecular mechanisms underlying regeneration with links to neuroscience, stem cell biology and neuropeptide structure and function in the absence of a centralised nervous system. 2) Comparative developmental events involving the evolution of biomineralization in the echinoderm clade. 3) Support for phylogenomic analysis in an otherwise little studied group. | ''Amphiura filiformis'' (O. F. Muller, 1776) is a brittle star found on the seabed of the European coasts from Greece to Norway and Sweden including the Baltic Sea . It is an important member of soft bottom communities where it digs a shallow burrow and feeds on plankton. It serves as food for crayfish and finfish. Stemming from its remarkable ability to regenerate arms in a matter of weeks it has become an emerging model for regeneration and stem cell biology in biomedical research. Research targets include: 1) The cellular and molecular mechanisms underlying regeneration with links to neuroscience, stem cell biology and neuropeptide structure and function in the absence of a centralised nervous system. 2) Comparative developmental events involving the evolution of biomineralization in the echinoderm clade. 3) Support for phylogenomic analysis in an otherwise little studied group. | ||
Development raw reads are available at NCBI accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA349786 PRJNA349786]. | Development raw reads are available at NCBI accession [https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA349786 PRJNA349786]. | ||
Revision as of 19:55, 3 February 2023
Future Species
Apostichopus parvimensis
The Warty Sea Cucumber (Apostichopus parvimensis) is a Sea Cucumber that can be found from the Gulf of Alaska to southern California. It is found from the low intertidal zone to a depth of 250 m. They are most abundant in areas with moderate current with cobbles, boulders or bedrock.
The Warty Sea Cucumber can grow to a length of 60 cm and a width of 5 cm. It has a soft, cylindrical body, with red-brown to yellowish leathery skin. There are numerous grey spots along its body, hence the name 'warty'. It has an endoskeleton just below the skin. The mouth and anus are on opposite sides of the body. The mouth is surrounded by ten retractable tentacles that are used to bring food in. Five rows of tube feet extend from the mouth to the anus. Mobility is limited, though individuals can move up to 4 m per day while feeding.
A. parvimensis is a solitary nocturnal animal. It has the ability to regenerate all parts of its body. When threatened, it can expel all its internal organs through its anus and grow new ones. It can also expel sticky filaments to ensnare or confuse predators. It undertakes seasonal migrations to different depths. These Sea Cucumbers have separate sexes, and eggs are fertilized externally. Spawning usually takes place in November, and each female can produce thousands of eggs. After fertilization, a larva is formed which metamorphoses into a Sea Cucumber after a few weeks.
The latest assembly is on NCBI with accession PRJNA182998
Ophiothrix spiculata
The Spiny Brittle star, Ophiothrix spiculata Le Conte, 1851, is a member of the Ophiuroidea another of the five classes of echinoderms. Brittle stars have a central disk that is clearly separated from long flexible arms often covered with spines. O. spiculata ranges from Moss Beach, California to Peru including the Galapagos Islands. It lives from the low rocky intertidal, under rocks, in crevices, and in algal holdfasts to subtidal depths of ~2,000 meters. Little is known about reproduction but spawning has been noted in July at Pacific Grove, California. Juveniles have been recovered from settling panels in the Monterey Bay from April to August.
The genome of O. spiculata is being sequenced by Baylor College of Medicine, Human Genome Sequencing Center under the NCBI project number PRJNA182997.
Eucidaris tribuloides
Eucidaris tribuloides, sometimes called the slate pencil urchin, belongs to the order Cidaroida. It is the sister group of the Euechinoida, the order which contains the more common sea urchins of North American shores such as S. purpuratus and L. variegatus.
These two branches of the echinoderm phylogenetic tree diverged about 255 MYA. E. tribuloides is found from North Carolina to Brazil and throughout the Caribbean Basin. Although it ranges down to 800 meters in depth is most commonly found from the surface to 50 m. The slate pencil urchin offers a research model with distinctly different developmental features to the usual models mentioned above. In particular it has a variable number of micromeres and a total lack of early ingressing mesenchyme, a group of cells that secretes the larval skeleton in other species.
The genome assembly is available on NCBI at accession PRJNA63057.
Mesocentrotus franciscanus
Red sea urchins, Mesocentrotus franciscanus (A. Agassiz, 1863) range from Alaska to Baja California at depths from just below the low tide line to 90 m. It prefers rocky shores that are sheltered from strong wave action. Although the red urchin has long been called Strongylocentrotus franciscanus, recent molecular phylogenies support the name to Mesocentrotus franciscanus (A. Agassiz, 1863) (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88).
A low coverage sequence of this species is available at NCBI at accession PRJNA20313.
Allocentrotus fragilis
Allocentrotus fragilis, the fragile sea urchin, lives on the sea floor at depths from 300 to 1600 ft. While Allocentrotus fragilis has long been the accepted name used for this species in the family Strongylocentrotidae, a more appropriate name might be Strongylocentrotus fragilis (Jackson, 1912). Indeed, the molecular evidence fully supports this change of name (Kober and Bernardi BMC Evolutionary Biology 2013, 13:88).
A low coverage sequence of this species is available at NCBI at accession PRJNA20317.
Amphiura filiformis
Amphiura filiformis (O. F. Muller, 1776) is a brittle star found on the seabed of the European coasts from Greece to Norway and Sweden including the Baltic Sea . It is an important member of soft bottom communities where it digs a shallow burrow and feeds on plankton. It serves as food for crayfish and finfish. Stemming from its remarkable ability to regenerate arms in a matter of weeks it has become an emerging model for regeneration and stem cell biology in biomedical research. Research targets include: 1) The cellular and molecular mechanisms underlying regeneration with links to neuroscience, stem cell biology and neuropeptide structure and function in the absence of a centralised nervous system. 2) Comparative developmental events involving the evolution of biomineralization in the echinoderm clade. 3) Support for phylogenomic analysis in an otherwise little studied group. Development raw reads are available at NCBI accession PRJNA349786.
Previous Assemblies by Species
Strongylocentrotus purpuratus
Assembly_3.1 (Spur_3.1)
README for Genome Sequence of Strongylocentrotus purpuratus, Spur_v3.1 (June 15th, 2011)
Conditions for Use
The data may be freely downloaded, used in analyses, and repackaged in databases. Some of the data presented here represents work inprogress. It is being released by the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) prior to project completion as a public service to allow our colleagues to search for genes or functions and speed their research. These data have not been edited and are presented "as is." You should regard the data as preliminary if it is unpublished. The data providers and associated funding agencies bear no responsibility for the user's reliance upon or interpretation of these data. The accuracy or reliability of the data is not guaranteed or warranted in any way and the providers disclaim liability of any kind. If you use this preliminary information we request that you honor the following conditions: Please communicate your results to us so that we can incorporate them into the annotation of the final sequence. Contact us at hgsc-help@hgsc.bcm.tmc.edu. Acknowledge the information obtained from BCM-HGSC in publications by stating in Materials and Methods and Acknowledgements: "Preliminary sequence data was obtained from Baylor College of Medicine Human Genome Sequencing Center website." Also acknowledge our funding source, which is listed in each project, with a statement such as "The DNA sequence of [organism] was supported by [grant number from funding agency to PI] at the BCM-HGSC." We also request that you notify us when your manuscript is accepted and send us a pre-print of the article. Use of this data or information derived from it on a web page is permitted, providing the web page contains the statement that "Preliminary sequence data was obtained from the Baylor College of Medicine Human Genome Sequencing Center website." Please inform us of your web page by sending email to hgsc-help@hgsc.bcm.tmc.edu. All other written or oral public disclosures of research using data from the BCM-HGSC should follow the acknowledgment guidelines outlined above. However, although we encourage use of this preliminary information for limited studies, we request that you do not publish whole genome or chromosome scale analyses of genes or genomic data prior to the publication of the BCM-HGSC report on the final genome sequence and analysis. Contact the BCM-HGSC at hgsc-help@hgsc.bcm.tmc.edu to discuss a waiver of this request, which could involve simple acknowledgment, co-authorship, or other methods. Any redistribution of the data should carry this notice.
What's New
This is the eighth release (Spur_3.1) of the genome assembly of sea urchin, Strongylocentrotus purpuratus. This assembly used additional Illumina reads with different end sequence spacing, known as a rainbow library series. The rainbow libraries consist of 4 libraries, a fragment paired end library with ~300bp insert size, and three mate-pair libraries with 1kb, 3kb and 5-6kb inserts. Each library has approximately 10x sequence coverage of genome (see Read statistics below for details). The reads were mapped to the Spur_v2.6 genome assembly and then were used for superscaffolding using the Atlas-Link software and local assembly and gap filling using Atlas-GapFill software. The scaffold N50 increased from 168kb (Spur_v2.6) to 404kb and the contig N50 increased by ~2kb.
Introduction
This is a draft assembly which may contain errors so users should exercise caution. Typical errors in draft genome sequences include misassemblies of repeat sequences, collapses of repeat regions, and artificial duplications in highly polymorphic regions. However base accuracy in contigs is usually very high with most errors near the ends of contigs. A rainbow library sequencing strategy was used to increase the contiguity of the S. purpuratus genome assembly, increasing the average scaffold length and closing assembly gaps. Four different shotgun libraries with nominal insert sizes of ~300bp, 1k, 3k, 5-6kb were constructed for Illumina sequencing. Reads from the recircularized 1k, 3k, 5-6kb libraries were trimmed from the 3' end to different lengths (50bp, 80bp, 120bp) and mapped. Reads that could be mapped were retained at the longest length that mapped to avoid the mapping issues created when the junction fragment is included in the mapped sequence.The reads from the shorter insert paired end library were mapped using the entire read length. After mapping, all reads were combined and used for super-scaffolding and intra-scaffold gap filling with the Atlas-link software. Then a local assembly of reads around each assembly gap was carried out to further fill the gaps using the Atlas-GapFill software. As a result, the scaffold N50increased to 404 Kb and the contig N50 increased by around 2kb. Comparison to the 17,461 available S. purpuratus Unigene sequences from NCBI showed the genome assembly is nearly complete (see the Sequence and Scaffold statistics section below). The number of Unigene contigs aligning 100% of their length increased by 0.1% .
Description of files
The files can be found on the HGSC ftp site ftp://ftp.hgsc.bcm.edu/Spurpuratus/fasta/Spur_v3.1/
- Contigs directory
- This directory has 3 files for assembled contigs in the genome, there are no chromosome assignments for the contigs in Spur_3.1. The .gz files are compressed with gzip.*Spur_3.1.AGP (AGP file)*Spur_3.1.contig.fa (contig fa)*Spur_3.1.contig.qual (contig quality)*acc_ctg_num.tbl (table listing GenBank accession number for each contig)The AGP file describes how to combine the individual contigs to create the linearized genome sequences in the LinearScaffolds directory.
- Linear Scaffolds directory
- This directory has 1 fasta file and 1 quality file compressed with gzip.
- Spur_3.1.linearScaffold.fa (scaffold linear scaffold sequence)
- Spur_3.1.linearScaffold.qual (scaffold linear scaffold sequence quality)The fasta formatted sequence files are for linearized scaffolds where the gaps between adjacent contigs within a scaffold are filled with 'N's and the captured gap size is estimated from the clone insert size. Each scaffold is a separate sequence within the files.
Sequence and Scaffold Statistics
| Assembly | Type | Number | N50 (kb) | Bases+Gaps (Mb) | Bases (Mb) |
|---|---|---|---|---|---|
| Spur_v3.0 | Scaffolds | 31,238 | 404,330 | 936,069,451 | 816,170,552 |
| Spur_v3.0 | Contigs | 174,512 | 13,472 | 816,170,552 | 816,170,552 |
Alignment of scaffolds to 17,461 Unigene contigs before and after upgrade
Percent of Scaffolds Aligning to Genome over Alignment Length
| Alignment length | 100% | 95% | 80% | 50% |
|---|---|---|---|---|
| Spur_v2.6 | 89.30% | 99.10% | 99.80% | 99.90% |
| Spur_v3.0 | 89.40% | 99.20% | 99.80% | 99.90% |
Read Statistics
| PE (300 bp) | 1 kb | 3 kb | 5-6 kb | |
|---|---|---|---|---|
| Total reads | 69,680,000 | 73,200,000 | 84,453,334 | 85,480,000 |
| Read length | 125bp | 150bp | 150bp | 150bp |
| Mapped | 67.0% | 67.2% | 68.2% | 64.4% |
| Bridge contigs | 3,840,891 | 6,600,203 | 9,684,914 | 10,558,589 |
| Within contigs | 32,467,846 | 21,503,082 | 20,154,150 | 14,136,572 |
| Mis-oriented [1] | 32,996 | 41,662 | 74,554 | 67,288 |
| Over distance [2] | 763,098 | 430,730 | 435,184 | 678,546 |
| Good pairs [3] | 31,671,752 | 21,030,690 | 19,994,412 | 13,390,738 |
[1] Mis-oriented reads map with an orientation of the two ends of the pair that is not expected. For PE, reads are expected to be oriented as -> .
[2] Over distance indicates the count of mates with excessive distance between mates, the following insert size cutoff were used:PE: >800bp1k: > 2000bp3k: > 4000bp5-6k: > 8000bp
[3] Good pairs refers to pairs in expected orientation and insert size.
Spur_3.1 (June, 2011)Contamination removed version of Spur_3.0.Spur_v3.0 (March, 2011)This release is an improved assembly using a variety of Illumina libraries with different mate-pair distances for scaffolding and gap filling. Spur_v2.6 (April, 2010)Contamination removed version of Spur_v2.5 Spur_v2.5 (February, 2010)Improved assembly of Spur_v2.1 using SOLiD mate pairs.Spur_v2.1 (September, 2006)This release is based on Spur_v2.0, with contaminations removed. Spur_v2.0 (June, 2006) This release is an independent assembly that combines BAC skim readsand WGS reads.Spur_v0.5 (April, 2005) This release update removed 716 contigs of contaminating(non-S. purpuratus) sequence and overlapping (second haplotype contigs).Otherwise the assembly statistics remain unchanged.Spur_v0.4 (March, 2005) ftp://ftp.hgsc.bcm.tmc.edu/pub/data/This release updated the agp file to omit scaffolds of contaminating(non-S. purpuratus) sequence and update coordinates for 65 pairs ofoverlapping contigs. Otherwise the assembly statistics remain unchanged.Spur_v0.3 (November, 2004)
This release is the first, preliminary assembly of the California purple sea urchin, Strongylocentrotus purpuratus, genome.
Assembly 2.6 (Spur 2.6)
This version of the updated Strongylocentrotus purpuratus genome sequence was derived from the Spur2.5 version through the removal of contaminating E. coli sequences. The gene sequences have changed very little.
Assembly_2.5 (Spur_2.5)
README for Genome Sequence of Strongylocentrotus purpuratus, Spur_2.5(February 11, 2010)
What's New
This is the release (Spur_v2.5) of the upgraded genome assembly of sea urchin Strongylocentrotus purpuratus using additional ABI SOLiD sequence for superscaffolding and gap filling. The scaffold N50 increased by 43kb and the contig N50 increased by 2kb.
Introduction
The draft assembly may contain errors so users should exercise caution. Typical errors in draft genome sequences include misassemblies of repeat sequences, collapses of repeat regions, and artificial duplications in highly polymorphic regions. However base accuracy in contigs is usually very high with most errors near the ends of contigs. Additional sequence coverage generated using the ABI SOLiD technology from small insert fragments was incorporated into this version of the assembly. The paired-end reads were used for superscaffolding and intra-scaffold gap filling. The assembly contains new scaffolds formed from merging scaffolds and scaffolds where some intra scaffold gaps are filled by other scaffold or contigs. As a result, the scaffold N50 increased to 166Kb and the contig N50 increased by 2kb. The additional sequence is ~18x genome coverage and the reads have an average insert size of ~1.5k. 500 million reads and have a read length of 25bp, and 46 million reads have a read length 50bp. Out of the total ~273 million clones, 30 million have both ends uniquely mapped. The 13% of these uniquely mapping pairs (4 million) that link two different scaffolds were used to upgrade the assembly with a recently developed scaffolding algorithm.Comparison to the 17,461 available S. purpuratus Unigene sequences from NCBI showed the genome assembly is nearly complete (see section 4). The number of Unigene contigs aligning over 95% or more of their length and the number of Unigene contigs aligning over 80% or more of their length both increased by 0.1% Comparison to the SOLiD sequencing reads (see read statistics section 5) confirmed the quality of the assembly. Over 99.8% of the pairs of uniquely mapping reads within scaffolds were correctly oriented. Over 99.6% of the pairs of uniquely mapping reads within scaffolds had insert sizes of
Description of files
The files can be found on the HGSC ftp site ftp://ftp.hgsc.bcm.edu/Spurpuratus/fasta/Spur_v2.5/ This directory has 3 files for assembled contigs in the genome, there are no chromosome assignments for the contigs in Spur_v2.5. The .gz files are compressed with gzip. Spur2.5.AGP (AGP file)Spur2.5.contig.fa(contig.fa)Spur2.5.contig.fa.qual (contig quality)
- Contigs/ directory
- The AGP file describes how to combine the individual contigs to create the linearized genome sequences in the LinearScaffolds directory.
- linearScaffolds/ directory This directory has 1 fasta files and 1 quality files compressed with gzip. Spur2.5.linearScaffold.fa (scaffoldlinear scaffold sequence) Spur2.5.linearScaffold.fa.qual (scaffold linear scaffold sequence quality) The fasta formatted sequence file (Spur2.5.AGP.linearScaffold.fa) are for linearized scaffolds where the gaps between adjacent contigs within a scaffold are filled with 'N's and the captured gap size is estimated from the clone insert size. Each scaffold is a separate sequence within the files.
- unassembled/ directory This directory has 2 files which have not changed from the previously assembly, Spur_v2.1 Spur_v2.1.unassembled.reads.fa.gz (unassembled reads fasta file) Spur_v2.1.unassembled.reads.fa.qual.gz (unassembled reads quality file). The unassembled reads files contain reads that are not used in the assembly.
Sequence and Scaffold statistics before and after upgrade
| Assembly | Type | Number | N50 (kb) | Bases+Gaps (Mb) | Bases (Mb) |
|---|---|---|---|---|---|
| Spur_2.5 | Scaffolds | 77,726 | 166,504 | 919,498,871 | 813,404,257 |
| Spur_2.1 | Scaffolds | 114,222 | 123,485 | 907,070,087 | 810,023,010 |
| Spur_2.5 | Contigs | 198,392 | 11,500 | 813,404,257 | 813,404,257 |
Alignment of scaffolds to Unigene* contigs before and after upgrade
Alignment of scaffolds to Unigene* contigs before and after upgrade
| Alignment length | 100% | 95% | 80% | 50% |
| Scaffolds Aligning** | 89.30% | 99.10% | 99.80% | 99.90% |
| Scaffolds Aligning*** | 89.30% | 99.20% | 99.90% | 99.90% |
- Total: 17,461 (**before ***after)
Unigene sequences used for completeness check.
Read Statistics
Sequence Coverage [6] 18x
| Total Raw reads (25bp) | 250,382,437 | 249,997,486 | 500,379,923 |
| Raw reads (50bp) | 43,267,758 | 43,279,703 | 86,547,461 |
| Uniquely mapped (25b)[1] | 27,879,092 | 27,879,092 | 55,758,184 |
| Uniquely mapped (50b)[1] | 3,912,119 | 3,912,119 | 7,824,238 |
| Bridge scaffolds (25bp)[2] | 3,323,043 | 3,323,043 | 6,646,086 |
| Bridge scaffolds (50bp)[2] | 720,919 | 720,919 | 1,441,838 |
| Within scaffold (25bp)[3] | 24,556,049 | 24,556,049 | 49,112,098 |
| Within scaffold (50bp)[3] | 3,191,200 | 3,191,200 | 6,382,400 |
| Mis-oriented reads (25bp)[4] | 33,671 | 33,671 | 67,342 |
| >5kb insert size (25bp)[5] | 94,060 | 94,060 | 188,120 |
[1] Reads from clones whose F3 end and R3 end both uniquely mapped.
[2] Reads which are from [1] and whose F3 end and R3 end are mapped to two different scaffolds.
[3] Reads which are from [1] and whose F3 end and R3 end are mapped to same scaffold.
[4] Reads which are from [3] 25bp and whose F3 end and R3 end are mapped in wrong orientation.
[5] Reads which are from [3] 25bp and whose inferred insert size from mapping are bigger than 5k, too big to be realistic.
[6] Sequence coverage was calculated as the total SOLiD reads bases divided by estimated genome size (800 Mb).
History Spur_v2.5 (Feburary, 2009)
Improved assembly of Spur_v2.1 using SOLiD mate pairs.Spur_v2.1 (September, 2006) This release is based on Spur_v2.0, with contaminations removed. Spur_v2.0 (June, 2006)
This release is an independent assembly that combines BAC skim readsand WGS reads.Spur_v0.5 (April, 2005)
This release update removed 716 contigs of contaminating (non-S. purpuratus) sequence and overlapping(second haplotype contigs). Otherwise the assembly statistics remain unchanged.Spur_v0.4 (March, 2005)
This release updated the agp file to omit scaffolds of contaminating (non-S. purpuratus) sequence and update coordinates for 65 pairs of overlapping contigs. Otherwise the assembly statistics remain unchanged.Spur_v0.3 (November, 2004)
This release is the first, preliminary assembly of the California purple sea urchin, Strongylocentrotus purpuratus, genome.
Assembly_2.1 (Spur_2.1)
Spur_2.1 combines BAC reads and WGS reads and utilizes BAC tiling path information. Contaminations identified in Spur_2.0 were removed. Compared to previous assembly releases, Spur_2.1 is more continuous and has fewer false duplications. The Spur_2.1 release was assembled from 2-fold average coverage in sequence reads from Bacterial Artificial Chromosomes (BAC) and 6-fold coverage in Whole Genome Shotgun (WGS) with the HGSC Atlas-2.0 genome assembly system at Baylor College of Medicine. The BAC reads were produced by the Clone-Array Pooled Shotgun Sequencing method (CAPSS) from BAC clones selected based on a minimal FingerPrinted Contigs (FPC) tiling path.In CAPSS pooled BAC reads are assigned to individual BACs by deconvolution. Each BAC assembly was enriched with WGS reads that overlap with the individual BAC reads. The mixed reads sets were assembled locally with Atlas. Sets of overlapping BAC clones were identified based on shared WGS reads and sequence overlaps. The overlapping enriched BACs were then merged together to form the backbone of genome assembly. The merged BAC assemblies were further scaffolded using information from mate pairs, BAC clone vector locations, and BAC tiling path information. Finally contigs from the WGS assembly Spur_0.5 were used to fill gaps in BAC assembly to produce Spur_2.0 release. Extensive contamination analysis was done on Spur_2.0 release. Spur_2.1 release was produced by removing contaminated sequences from Spur_v2.0 release. The Spur_2.1 release includes a set of contigs (continuous blocks of sequence) and scaffolds. Scaffolds include sequence contigs that can be ordered and oriented with respect to each other (multi-contig scaffolds) as well as contigs that could not be linked (single-contig scaffolds or singletons). The N50 of the scaffolds associated with BACs is 216 Kb.The N50 of all scaffolds is 142 Kb. The total length of all contigs greater than 1kb is 804 Mbps. When the gaps between contigs in scaffolds are included, the total pan of the assembly is 907 Mbps. The estimated size of the genome based on the assembly is 814 Mbps.The Spur_2.1 assembly was compared with other available sea urchin sequence data (ESTs, Unigene clusters) to determine the extent of coverage (completeness). A preliminary examination showed over 90% of the sequences in this data set is represented, indicating that the shotgun libraries used to sequence the genome were comprehensive. Typical errors in draft genome sequences include misassemblies of repeat sequences, collapses of repeat regions, and artificial duplications in polymorphic regions. However base accuracy in contigs is usually very high with most errors near the ends of contigs. These data can be downloaded from
ftp://ftp.hgsc.bcm.edu/Spurpuratus/fasta/Spur_v2.1/
Assembly_0.5 (Spur_0.5)
Spur_0.5 is a preliminary assembly of the California purple sea urchin, S. purpuratus, using whole genome shotgun(WGS) reads with the Atlas genome assembly system at the Baylor college of Medicine Human Genome Sequencing Center.The products of the Atlas assembler are a set of contigs and scaffolds. The total length of all contigs greater than 1kb is 768Mb, the N50 of the contigs larger than 1kb is 10.18 kb and the N50 of the scaffolds is 47.98 kb. The total span of the assembly is 1.13 Gb, which is 240 Mb larger than the estimated genome size. The sequence coverage is 6.2X.A preliminary examination showed that over 90% of the sequences in other available sea urchin sequence data sets (Unigene clusters) is represented in the Spur_0.5 assembly. By comparison to 25 NCBI HTGS_PHASE2 BACs( total 2.9Mb), some types of inconsistency were found: several cases of short non-merging overlaps were observed, most at the tail of scalffolded contigs. this may due to polymorphism such that the merging criteria were not met.several short contigs were found aligning in the middle of long alignment gaps of large scaffolded contigs (7 cases), these large gaps come from scaffolding with only short (2 ~6k) and large (50k, 150k)inserts but no middle sized (10 ~ 15 k) inserts, resulting in unfilled large gaps and artificial expansion of total sequence size in the super contigs. Other minor inconsistencies included three cases of differences between genome contigs and PHASE2 BACS, and two possible misjoins. Checking the three contigs in detail did not identify misassemblies. One possible misjoin is in a repeat region and one is a possible local misordering of a short 2k contig in the middle long scaffolded contig.
Difference between 0.5, 2.0 and 2.1
Introduction
All analysis published in sea urchin genome paper were based on 0.5 assembly version of the genome. Subsequently, Baylor released 2.0 and 2.1 assembly versions. Substantial improvements were achieved from 0.5 to 2.0, whereas the 2.1 assembly was a cleaned up version of 2.0.
Baylor's description of 2.0 and 2.1 assemblies
Baylor released the following comments to describe 2.0 and 2.1 assemblies:
Spur_2.1 combines BAC reads and WGS reads and utilizes BAC tiling path information. Contaminations identified in Spur_2.0 were removed. Compared to previous assembly releases, Spur_2.1 is more continuous and has fewer false duplications. The Spur_2.1 release was assembled from 2-fold average coverage in sequence reads from Bacterial Artificial Chromosomes (BAC) and 6-fold coverage in Whole Genome Shotgun (WGS) with the HGSC Atlas-2.0 genome assembly system at Baylor College of Medicine. The BAC reads were produced by the Clone-Array Pooled Shotgun Sequencing method (CAPSS) from BAC clones selected based on a minimal Finger Printed Contigs (FPC) tiling path. In CAPSS pooled BAC reads are assigned to individual BACs by deconvolution. Each BAC assembly was enriched with WGS reads that overlap with the individual BAC reads. The mixed reads sets were assembled locally with Atlas. Sets of overlapping BAC clones were identified based on shared WGS reads and sequence overlaps. The overlapping enriched BACs were then merged together to form the backbone of genome assembly. The merged BAC assemblies were further scaffolded using information from mate pairs, BAC clone vector locations, and BAC tiling path information. Finally contigs from the WGS assembly Spur_0.5 were used to fill gaps in BAC assembly to produce Spur_2.0 release. Extensive contamination analysis was done on Spur_2.0 release. Spur_2.1 release was produced by removing contaminated sequences from Spur_v2.0 release.
Comparison between 2.0 and 2.1
Because 2.0 assembly is not much different from its better version 2.1, most of our bioinformatics calculations are being done on 2.1. We only made quick comparison between 2.0 and 2.1 and found them to be nearly identical. Among 114224 scaffolds of 2.1 assembly, 113694 were fully copied from 2.0 and 530 were different. Among those 530 scaffolds in 2.1, 249 + 9 were parts of 2.0 scaffolds, whereas 272 were of same length as 2.0 version but with N regions filled up.
Comparison between 0.5 and 2.1
We generated complete maps between V0.5 and V2.1 genomes. These maps can be used to convert any previously developed resources on V0.5 to V2.1 assembly.Among all 114222 V2.1 scaffolds, 83754 are identical to V0.5 scaffolds. The remaining ~30K scaffolds of V2.1 assembly are significantly different from V0.5. Most of them are large scaffolds containing most SPU genes. 4026 are super-sets of 6572 V0.5 scaffolds and 6106 are part of V0.5 scaffolds. For the last case, V0.5 assembled scaffolds were incorrect and broken into parts.
Mapping procedure
The maps were generated in the following manner.
- All 30 mers in the entire genomes of 0.5 and 2.1 assemblies were determined.
- Those sequences were binned together and only the ones satisfying the following criteria were kept: (a) 30-mer matched W strand of 0.5 genome, (b) 30-mer had exactly one match each in 0.5 and in 2.1 genomes.
- Those unique 30-mers were combined into longer overlapping regions between the genomes. Because of the way the 30-mers were screened, the derived regions are unambiguous - i.e. repetitive regions are not expected to create any duplication in the mapping.
- Neighboring fragments from the genome were combined into longer identical genomic regions.The created overlap file can be used to map any region in one assembly to another, unless the segment is on a repeat region and cannot be uniquely mapped to the other genome.
SpPtBB Genome Assembly
Introduction
The SpPtBB assembly uses the same data sets employed by Spur4.2 plus an additional Illumina data set (100x coverage, 400bp insert, paired ends). Since the assemblies are independent, regions where SpPtBB and Spur4.2 agree are more likely to be correct, and regions where they differ more likely to be in error (in one or both). The process consists of many steps whose commands are presented in some detail in SpPtBB_commands.txt. A summary of the steps is presented below. The method is very similar to that used for the LvPtE5C assembly.
Illumina Read Contamination
The new S. purpuratus Illumina reads were found to be contaminated with Mus musculus mRNA sequences like NM_018794 and NM_010511. Corresponding sequences should not be present in the later linkage steps employing Sanger and PacBIO sequences, so the mammalian sequence contaminants should primarily appear in small scaffolds.
Platanus Production of Contigs
Platanus is used to produce an initial set of contigs from the 100X Illumina paired end data set. This initial set of contigs often contains for any genome region copies from both haplotypes. This was true even though the -u 1.0 command line parameter was used, which should merge some of these haplotype pairs into a single sequence. Because of the highly polymorphic nature of this genome the common deduplication tools are not very good at identifying and eliminating one sequence from each such pair. This is especially true because such sequences often overlap with an offset, which the deduplication tool employed would not recognize as a duplication. Each time two sequences are joined and the gap filled that provides another opportunity for the deduplication method to remove one half of such a pair. This process is repeated three times below.
Assembly Overview
- Platanus trim 100X Illumina PE data (SRR7211988)
- Platanus trim Illumina MP data (SRR446979,SRR446980,SRR446981)
- Platanus assemble trimmed 100X Illumina PE data
- Gather Sanger reads into files: all unmatched read to single_for (with its reverse complement as single_rev), then into _for/_rev files by documented insert size in kbp of 2, 3, 3.5, 4.5, 5, and 6. Some ambiguously documented pairs used insert size of 1999bp.
- Platanus scaffold contigs using 100X Illumina PE, Illumina MP, all Sanger pairs.
- Platanus gap_close using 100X Illumina PE, Illumina MP, all Sanger pairs.
- FGAP (in a wrapper script) to fill gaps with megareads (from a MaSuRCA run using 100X Illumina PE and PacBIO reads).
- bbmap dedupe.sh to remove duplicates
- Map Sanger and Illumina mate pair reads onto dedup sequences using Opera preprocess_reads.pl, makes BAM files
- OPERA-pacbio-read.pl to prepare more BAM and other linkage files from ccs corrected PacBIO reads and 100x Illumina paired end data onto dedup sequences.
- OPERA-LG to scaffold
- FGAP (in a wrapper script) to fill gaps with megareads.
- bbmap dedupe.sh to remove duplicates
- P_RNA_scaffolder to scaffold using RNA-seq reads same as SRR532151 (Illumina RNA-seq)
- FGAP (in a wrapper script) to fill gaps with megareads.
- bbmap dedupe.sh to remove duplicates
Sequence Statistics
Sequence statistics are for the final scaffold SpPtBB.fa.
| Scaffolds/Contigs | Number | N50 (kb) | Bases (Mb) | Gap (Mb) |
|---|---|---|---|---|
| All Scaffolds | 494117 | 67.11 | 919.5 | 67.41 |
| All Contigs | N/A | N/A | N/A | N/A |
Haplotype Specific Contigs
Introduction
Previous assemblies have used methods which attempt to collapse both haplotypes into a single representation of the genome. Because the organism is roughly 1% polymorphic, and much of that is in the form of indels, the resulting "average" sequence typically corresponds to neither of the original haplotypes. The resulting mixing may cause experiments to fail, for instance PCR runs may not work when the mixed sequence is used to design the primers, even when the DNA from the reference organism is used. Consequently, an attempt was made to assemble the genome while keeping the two haplotypes separate.
Method
Tuples to sticks
A 100X coverage Illumina paired end data set was obtained with 150bp reads and 400bp inserts. Using sticker and other locally written software the properties of 23bp tuples derived from these reads were analyzed. The number of instances of each unique set of tuples {A,B,C} was determined, where A is the tuple shifted 1 bp 5' from B, C is the tuple shifted 1 bp 3' from B. These are stored in their canonical forms along with the relative orientation of A and C to B. From this data "sticks" were determined, where a stick is a sequence of tuples where the count varies only a little at a time and the path to other tuples does not fork. Sticks of 1 bp length were reclassified as "complex" if there did not exist a 1:1 mapping between the observed and 5' and 3' bases. (That is if only A->C and C->T were observed it remained a stick, whereas A->C, C->T, and C->G became a "complex", since observing C at the 5' end did not predict a single base at the 3' end.) An ad hoc algorithm was then applied to determine the estimated ploidy of each stick. This was essentially a check of mass conservation - the ploidy of a stick which forked into 2 other sticks at each end must be the sum of the ploidies of those sticks. Sticks with ploidies above 8 by their raw counts were locked at those values. This determined a set of 1X sticks to serve as unitigs. (In a typical diploid assembly the unitigs correspond to what are described here as 2X sticks.)
Sanger Read Preprocessing
The Sanger reads used in the other assemblies were then used to provide linkage between the 1X sticks. These reads were quality filtered using Lucy. During the course of this work it was discovered that many templates contained false palindromes. These are the result of contamination of the sequencing primers, so that both the forward and reverse primers were present in a reaction, with one at a much lower concentration than the other. When the more prevalent primer reached a difficult sequence the amplitude would be drastically reduced, revealing the sequence from the other primer, which then became the dominant sequence. The result of this would be that from the nth base on both the forward and reverse sequences from that template would be the same. A Smith-Waterman alignment was made from the forward and reverse alignments for each template that had both using ssw_test (a modified form of the test program from SSWlib), and those with a strong alignment on or very near the diagonal were clipped at the most 5' end of the alignment. In theory it should be possible to deconvolute the traces to determine which read is the rightful owner of the common sequence section, but no attempt was made to do so.
Illumina Read Contamination
Examination of the longest 1X sticks resulted in the discovery that some had very low tuple counts along their lengths. These were then found to correspond to Mus musculus mRNA sequences like NM_018794 and NM_010511. Evidently contamination occured at some point. Ideally another uncontaminated sequencing run would have been obtained, but that was not an option at the time. The Sanger reads were checked for several of the identified contaminants and these were not found. Because the linking step described below only connects sticks which have corresponding Sanger sequences it was hoped that this would eliminate most of the contaminants, and when the final contigs were checked, that was what was observed. However, the contaminants undoubtedly degraded some parts of the assembly, because a region which would otherwise have had a simple diploid "ladder" topology with two "rails" were complicated by a low intensity third "rail" from the contaminant.
Sanger Read Linking of Sticks
The Sanger reads were then used to build contigs from the previously identified 1X sticks. The trimmed and clipped Sanger reads were used to generate {template, canonical tuple, orientation, direction} objects. Linkage between pairs of 1X sticks based on these objects was used to generate contigs. Usually the linkage was direct, with several templates connecting a pair of sticks. In some instances linkage was inferred. Where two pairs of 1X sticks (A,a and B,b) flank a 2X stick, and the Sanger data linked A to B, then a was linked to b by elimination. For simple parts of the graph the sequence of intervening sticks was used to generate the contig sequence. For long or complicated intervening regions the sequence was determined using MAFFT to make a multiple sequence alignment of the Sanger sequences and a synthesized structure having the first 1X stick, a gap filled with N bases, and then the final 1X stick. The consensus of this alignment was then determined using fasta_consensus and used to fill the gap.
Results, Limitations, and Uses
The final contig set of 472178 members covers 1009448699 bp and has an N50 of 4804. Most contigs overlap with those from the other haplotype. Contigs from gene families or other very similar sequences with a relatively small number of copies are common. It is often difficult to determine in these cases which contig is a paralog and which is the other haplotype. Long repeat sequences are underrepresented because they may not contain any 1X sticks within a typical Sanger read length of another. Both the Sanger and Illumina reads are observed to fail consistently on certain genomic sequences which are thought to be "toxic" to PCR, and consequently this contig set contains few of those regions. When the contigs are mapped onto BACs constructed by other means pairs of these contigs are often found to flank one of these "toxic" regions, leaving a gap of several hundred base pairs.
The sequences present in this assembly are the best available representation of the haplotype variation in this genome and should be valuable when planning PCR and other related experiments which are critically sequence dependent. It must be emphasized that the difference between the two haplotypes of the sequenced individual is in no way special within this species. If a region of interest is found to differ greatly between these two haplotypes it would be safest to assume that that level of variation is present in other individuals of this species. Conversely, a strongly conserved region is likely well conserved in other individuals too. The contigs may be mapped onto a genomic scaffold using Blastn or a similar program. When doing so be aware that because the scaffold sequences are all a mixture of the two haplotypes the series of best matching haplotype specific contigs which results will be mixed as well - in such a map the haplotype may change between any pair of adjacent haplotype specific contigs.
Repeats (from SpPtBB)
Repeats for assembly SpPtBB were derived using the exact same method as for Lv repeats. There were 3579 sequences comprising 1847131 bp ranging in size from 33 bp to 9344 bp. These statistics are very similar to those for Lytechinus variegatus.
Patiria miniata
V2.0 Assembly
We sought to improve the Patiria miniata genome assembly with additional PacBio sequences. We generated a new PacBio read dataset at the Duke University Sequencing Center using our reference individual DNA. The read dataset contains 2 million reads and 15.8 billion bp. The read N50 is 10.4 Kb. We used PBJelly2 to combine the PacBio reads with the previously assembled contigs. The results were an improvement in contig size and number with only a small reduction in the number of scaffolds (Table). The P. miniata Gene v2.0 set was generated using MAKER2 pipeline from v2.0 genome assembly.
| Pm v1.0 | Pm v2.0 | |
|---|---|---|
| Scaffold number | 60,183 | 57,698 |
| Scaffold N50 | 52,6141 | 76,341 |
| Contig number | 179,756 | 131,779 |
| Contig N50 | 9,466 | 18,676 |
V1.0 Assembly
What's New
Pmin_1.0 is the latest (as of Apr 11, 2012) assembly of the genome of Patiria Miniata. The assembly tools CABOG (Celera Assembler), Newbler, ATLAS-Link, and ATLAS-GapFill were used to assemble a combination of 454 reads (fragment and 2.5kb insert paired ends;~15x coverage) and Illumina reads (300bp insert and 2.5kb insert paired ends;~70x coverage).
Introduction
This information is for the first release (Pmin_1.0) of the draft genome sequence of the Patiria miniata . This is a draft sequence and may contain errors so users should exercise caution.Typical errors in draft genome sequences include misassemblies of repeated sequences, collapses of repeated regions, and unmerged overlaps(e.g. due to polymorphisms) creating artificial duplications.
With a goal of solving the polymorphism issues of the data while maintaining the sequence continuity, The Pmin_1.0 assembly was generated in the following steps:
- 454 reads were assembled by CABOG using settings less strignent than the default (unitigger=bog utgErrorRate=0.03 ovlErrorRate=0.08 cnsErrorRate=0.08 cgwErrorRate=0.14 doExtendClearRanges=0)
- Both contig and degenerate sequences from the previous step were chopped into fake reads with ~11x coverage (500bp long; 460bp overlap; 80bp minimal length) for ctgs and 8x coverage(450bp long; 400bp overlap; 80bp minimal length) for degs. The fake reads were then assembled by Newbler with the option of -large.
- Both 454 and iIlumina pair end reads were mapped to the contigs from the previous step. We used BLAT to map the 454 data and bwa(aln+samse) to map the Illumina data, both with the default options. Based on the mapping locations of the paired ends, contigs were then ordered and oriented into scaffolds using ATLAS-Link.
- ATLAS-GapFill was then used to assemble the reads locally in an attempt to fill the gaps among the contigs within the scaffolds.This final step produced 770.5Mb sequences with contig N50 size of 9.5kb and scaffold N50 size of 50.3kb.
Conditions for use
These data are made available before scientific publication with the following understanding:
- The data may be freely downloaded, used in analyses, and repackaged in databases.
- Users are free to use the data in scientific papers analyzing particular genes and regions if the providers of this data (Baylor College of Medicine Human Genome Sequencing Center) are properly acknowledged. Please cite the BCM-HGSC web site or publications from BCM-HGSC referring to the genome sequence.
- The BCM-HGSC plans to publish the assembly and genomic annotation of the dataset, including large-scale identification of regions of evolutionary conservation and other features.
- This is in accordance with, and with the understandings in the Fort Lauderdale meeting discussing Community Resource Projects and the resulting NHGRI policy statement (http://www.genome.gov/page.cfm?pageID=10506537).
- Any redistribution of the data should carry this notice.
Description of files
There are 2 directories.
- Contigs/ directory This directory has 2 files for assembled contigs in the genome, there is no chromosome assignment for the contigs in Pmin_1.0. Pmin_1.0.20120411.contigs.agp (agp file) Pmin_1.0.20120411.contigs.fa (fasta file) The Pmin_1.0.20120411.contigs.agp file describes the positions and orientations of the contigs in the group. It takes the standard NCBI format.
- LinearScaffolds/ directory This directory has 1 file Pmin_1.0.20120411.linear.fa The sequences are linearized scaffolds where the gaps between adjacent contigs within a scaffold are filled with 'N's and the captured gap size is estimated from the clone insert size.
Sequence Statistics
| Scaffolds/Contigs | Number | N50 (kb) | Bases (Mb) | Gap (Mb) |
|---|---|---|---|---|
| All Scaffolds | 60,336 | 50.3 | 811.6 | 41.1 |
| All Contigs | 181,436 | 9.5 | 770.5 | N/A |
History
Pmin_1.0 (Apr, 2012) This release was the first assembly of the Patiria Miniata genome.
Lytechinus variegatus
Assembly LvPtE5C
Introduction
The LvPtE5C assembly uses the same data sets employed by LvMSCB. In addition it uses the megareads from the LvMSCB assembly to scaffold and fill. LvPtE5C is not a derivative of either Lvar 0.4 or Lvar 2.2. Since the assemblies are independent, regions where LvPtE5C and Lvar 2.2 agree are more likely to be correct, and regions where they differ more likely to be in error (in one or both). The process consists of many steps whose commands are presented in some detail in LvPtE5C_commands.txt. A summary of the steps is presented below.
Illumina read contamination
The S. purpuratus Illumina reads produced at the same time were found to be contaminated with Mus musculus mRNA sequences like NM_018794 and NM_010511. The L. variegatus Illumina data appears not to be similarly contaminated, as these sequences were not found.
Platanus production of contigs
Platanus is used to produce an initial set of contigs from the 100X Illumina paired end data set. This initial set of contigs often contains for any genome region copies from both haplotypes. This was true even though the -u 1.0 command line parameter was used, which should merge some of these haplotype pairs into a single sequence. Because of the highly polymorphic nature of this genome the common deduplication tools are not very good at identifying and eliminating one sequence from each such pair. This is especially true because such sequences often overlap with an offset, which the deduplication tool employed would not recognize as a duplication. Each time two sequences are joined and the gap filled that provides another opportunity for the deduplication method to remove one half of such a pair. This process is repeated three times below.
Assembly overview
- Platanus trim 100X Illumina PE data (SRR7207203)
- Platanus trim Illumina MP data (SRR176809, SRR176810, SRR176812)
- Platanus assemble trimmed 100X Illumina PE data
- Platanus scaffold contigs using Illumina and 454 pair data.
- FGAP (in a wrapper script) to fill gaps with megareads.
- bbmap dedupe.sh to remove duplicates
- Map 454 and Illumina mate pair reads onto dedup sequences using Opera preprocess_reads.pl, makes BAM files
- OPERA-pacbio-read.pl to prepare more BAM and other linkage files from PacBIO reads and 100x Illumina paired end data onto dedup sequences.
- OPERA-LG to scaffold
- FGAP (in a wrapper script) to fill gaps with megareads.
- bbmap dedupe.sh to remove duplicates
- P_RNA_scaffolder to scaffold using RNA-seq reads in SRR1661111 (Illumina RNA-seq)
- FGAP (in a wrapper script) to fill gaps with megareads.
- bbmap dedupe.sh to remove duplicates
Sequence statistics
Sequence statistics for the final LvPtE5C file.
| Scaffolds/Contigs | Number | N50(kb) | Bases(Mb) | Gap(Mb) |
| All Scaffolds | 175668 | 55.11 | 938.8 | 33.19 |
| All Contigs | N/A | N/A | N/A | N/A |
Assembly LvMSCB
Introduction
The LvMSCB genome assembly was constructed with MaSuRCA using the 23x coverage 454 reads and 13x PacBIO reads previously employed in the Lvar 0.4 and 2.2 assemblies. In addition it used a new Illumina data set (100x coverage, 400bp insert, paired ends). This assembly has a large number of false joins, resulting from overly aggressive splicing through repeat sequences. However, for that reason it was very useful in determining the repeat sequences present in this genome. That is, long repeat sequences were created by overlapping very similar copies of that repeat, producing a longer repeat than could be otherwise built. However in doing so distant genomic regions were often joined across a hybrid repeat. Additionally, the superreads and megareads it produces are reasonably accurate representations of small sections of the genome. Contiguous nonrepetitive genome regions may be used to confirm predictions made in other assemblies but should not otherwise be relied upon.
Production of Superreads and Megareads
MaSuRCA's generation of megareads was slightly modified. The PacBIO reads were first passed through the CCS routine from Proovread to form, where possible, circular consensus sequences (see below). MaSuRCA then generated superreads from the 100x Illumina PE data. Each superread is a nonforking assembly of Illumina reads. These are mostly if not entirely haplotype specific. The Megaread production was modified slightly by adding Proovread's Siamaera detecting code, which is supposed to detect and truncate readthroughs of the SMRTbell adapter (see below). Megareads are PacBIO reads corrected using the superreads. These are not haplotype specific because the noisy nature of PacBIO reads often results in the best match being to a superread of the other haplotype. In other words, sections of Megareads are frequently converted from one haplotype to the other.
454 read processing
A file SRR.desc containing the list of all 40,454 SRA files for biosample SAMN00205415 was constructed, marking each SRR* with sn, pe, or rs (single, paired end, or RNA-seq). The sra files which were not rs were retrieved to a directory. These were processed as follows (extract and execinput are from drm_tools):
ls -1 *sra \
| extract -mt -dl '.' -fmt 'sffToCA -libraryname [1] -trim chop -output [1] [1].sff &'\
| nice execinput > sff2CAB.log 2>&1
- wait until all processes complete
cat SRR*frg >r454_all.frg
rm *fastq SRR*frg SRR*sra
Note, the preceding is not ideal because some of the SRR are from the same library and there may be duplicate reads in different files. For LvPtE5C sffToCA was rerun as follows:
export LIST=`extract -in SRR.desc -if ' sn ' -ifonly -mt -dl ' ' -fmt '[1].sff ' -eol -fileeol '\n'`
$TOPDIR/MaSuRCA/CA8/Linux-amd64/bin/sffToCA -libraryname single \
-trim chop -output single $LIST \
>sff2CA3S.log 2>&1
rm single.1.fastq single.2.fastq #these are empty
mv single.u.fastq single.fastq
export LIST=`extract -in SRR.desc -if ' pe ' -ifonly -mt -dl ' ' -fmt '[1].sff ' -eol -fileeol '\n'`
$TOPDIR/MaSuRCA/CA8/Linux-amd64/bin/sffToCA -insertsize 2500 250 -libraryname pairs \
-trim chop -linker titanium -output pairs $LIST \
>sff2CA3P.log 2>&1 &
#made pairs.1.fastq pairs.2.fastq pairs.u.fastq
A better cumulative frg file would have been:
cat single.frag pairs.frg >r454_all.frg
MaSuRCA configuration file
The MaSuRCA project.cfg contains:
DATA
PE= pe 400 20 $TOPDIR/Lv/Lv_ILLUMINA/pe_400_R1.fastq $TOPDIR/Lv/Lv_ILLUMINA/pe_400_R2.fastq
PACBIO=$TOPDIR/Lv/Lv_PACBIO/pacbio_all_CCS.fasta
OTHER=$TOPDIR/Lv/Lv_454/r454_all.frg
END
PARAMETERS
GRAPH_KMER_SIZE = auto
USE_LINKING_MATES = 1
LIMIT_JUMP_COVERAGE = 300
CA_PARAMETERS = cgwErrorRate=0.15 doExtendClearRanges=0
KMER_COUNT_THRESHOLD = 1
NUM_THREADS = 40
JF_SIZE = 80000000000
SOAP_ASSEMBLY=0
END
ccs preprocessing of PacBIO sequences
cd $TOPDIR/Lv/Lv_PACBIO
export PATH=$PATH:$TOPDIR/src/proovread/bin:$TOPDIR/src/proovread/util/bwa
export PATH=$PATH:$TOPDIR/src/proovread/util/Seq*/bin
export PERL5LIB=$TOPDIR/src/proovread/lib
fasta_to_fastq.pl
| ccseq -t 40 > out_ccs.fastq 2>out_ccs.log
#[17-08-15 11:43:24] [ccseq] Reading STDIN
#[17-08-16 09:37:14] [ccseq] Processed 6564119 subreads from 3060427 reads
#[17-08-16 09:37:14] [ccseq] 1403356 consensus + 1657071 bypassed single subreads
fastasplitn -in out_ccs.fastq -q4 -q2f -n 1 -p 1 \
| extract -if '>' -mt -dl ' ' -fmt '[1]' -wl 100000 >pacbio_all_CCS.fasta
mega_reads_assemble_nomatch_PROOVREAD.sh
The MaSuRCA provided mega_reads_assemble_nomatch.sh script modified to perform Proovread Siamaera processing is here: mega_reads_assemble_nomatch_PROOVREAD.sh.
assemble.sh scipt
The assemble.sh script produced by MaSuRCA was modified before running so that its last line became:
$TOPDIR/MaSuRCA/bin/mega_reads_assemble_nomatch_PROOVREAD.sh -t 40 \
-e $ESTIMATED_GENOME_SIZE -m work1 -p $TOPDIR/Lv/Lv_PACBIO/pacbio_all_CCS.fasta\
-a $TOPDIR/MaSuRCA/bin/../CA8/Linux-amd64/bin \
-o " $TOPDIR/Lv/Lv_454/r454_all.frg pe.linking.frg cgwErrorRate=0.15 doExtendClearRanges=0"
-B 17 -D 0.029
Sequence statistics
A deduplication step is applied to the scaffold file produced by the Celera Assembler to produce the final scaffold LvMSCB, whose statistics are shown below. The final contig file produced by the Celera Assembler whose statistics are shown here has not been similarly treated.
| Scaffolds/Contigs | Number | N50(kb) | Bases(Mb) | Gap(Mb) |
|---|---|---|---|---|
| All Scaffolds | 32916 | 59.47 | 1021 | 6.02 |
| All Contigs | 64833 | 30803 | 1058 | 1380 |
Assembly 2.2 (Lvar_2.2)
Assembly details, to the extent known, are described at the NCBI genome entry: GCA_000239495.2.
PacBIO sequence with 13x coverage, and the preceding Lvar 0.4 assembly, were processed with PBJelly to make new scaffolds.
Sequence statistics
| Scaffolds/Contigs | Number | N50(kb) | Bases(Mb) | Gap(Mb) |
|---|---|---|---|---|
| All Scaffolds | 322,794 | 46.35 | 1061 | 57.1 |
| All Contigs | 452,418 | 9.66 | 1004 | N/A |
Assembly 0.4 (Lvar_0.4)
Introduction
This information is for the first release (Lvar_0.4, NCBI GCA_000239495.1) of the draft genome sequence of the green sea urchin, Lytechinus variegatus . This is a draft sequence and may contain errors so users should exercise caution. Typical errors in draft genome sequences include misassemblies of repeated sequences, collapses of repeated regions, and unmerged overlaps (e.g. due to polymorphisms) creating artificial duplications. With a goal of solving the polymorphism issues of the data while maintaining the sequence continuity, The Lvar_0.4 assembly was generated in the following steps:
- 454 reads (fragment and 2.5kb insert pair ends;~13x coverage) were assembled by CABOG (Celera Assembler) using settings less stringent than the default (unitigger=bog utgErrorRate=0.03 ovlErrorRate=0.08 cnsErrorRate=0.08 cgwErrorRate=0.14 doExtendClearRanges=1) This step produced 716Mb of contig and 429Mb of degenerate sequences.
- Both contig and degenerate sequences from the previous step (a total of 1.1Gb) were chopped into fake reads with ~10x coverage (500bp long;450bp overlap;100 bp minimal length). The fake reads were then assembled by Newbler with the option of -large. This step produced 801Mb contig sequences with N50 size of 1.87kb, which was used as the backbone for the following process.
- Both 454 and iIlumina pair end reads (300 bp insert) were mapped to the contigs from the previous step. We used BLAT to map the 454 data and bwa(aln+samse) to map the Illumina data, both with the default options. Based on the mapping locations of the pair ends (2.5 kbp insert), contigs were then ordered and oriented into scaffolds using ATLAS-Link. Sum coverage for all Illumina reads was ~21x.
- ATLAS-GapFill was then used to assemble the reads locally in an attempt to fill the gaps among the contigs within the scaffolds. This final step produced 835Mb sequences with contig N50 size of 6.05kb and scaffold N50 size of 39.17kb.
Conditions for use
These data are made available before scientific publication with the following understanding:
- The data may be freely downloaded, used in analyses, and repackaged in databases.
- Users are free to use the data in scientific papers analyzing particular genes and regions if the providers of this data (Baylor College of Medicine Human Genome Sequencing Center) are properly acknowledged. Please cite the BCM-HGSC web site or publications from BCM-HGSC referring to the genome sequence.
- The BCM-HGSC plans to publish the assembly and genomic annotation of the dataset, including large-scale identification of regions of evolutionary conservation and other features.
- This is in accordance with, and with the understandings in the Fort Lauderdale meeting discussing Community Resource Projects and the resulting NHGRI policy statement (http://www.genome.gov/page.cfm?pageID=10506537).
- Any redistribution of the data should carry this notice.
Description of files
There are 2 directories.
- Contigs/ directory This directory has 3 files for assembled contigs in the genome, there is no chromosome assignment for the contigs in Lvar_0.4. Lvar_0.4.20110428.contigs.agp (agp file) Lvar_0.4.20110428.contigs.fa (fasta file) Lvar_0.4.20110428.contigs.fa.qual (qual file) The Lvar_0.4.20110428.contigs.agp file describes the positions and orientations of the contigs in the group. It takes the standard NCBI format.
- LinearScaffolds/ directory This directory has 2 files Lvar_0.4.20110428.linear.fa Lvar_0.4.20110428.linear.fa.qual The sequences are linearized scaffolds where the gaps between adjacent contigs within a scaffold are filled with 'N's and the captured gap size is estimated from the clone insert size.
Sequence Statistics
| Scaffolds/Contigs | Number | N50 (kb) | Bases (Mb) | Gap (Mb) |
|---|---|---|---|---|
| All Scaffolds | 330,611 | 39.17 | 966 | 131 |
| All Contigs | 518,238 | 6.05 | 835 | N/A |
Comparison to ESTs
The Lvar_0.4 assembly was compared to the 454 RNAseq assembly using BLAT: Align_Span/Isotig_Length
| >=50% | >=80% | >=95% | 100% | |
|---|---|---|---|---|
| a Percent | 99.5% | 97.8% | 93.4% | 69.2% |
| b Percent | 99.5% | 97.3% | 91.8% | 67.6% |
- After_Gast EST isotigs
- Before_Gast EST isotigs
History
Lvar_0.4 (Apr, 2011) This release was the first assembly of the Lytechinus variegatus genome.
Other Comments
Linkage limitations
Both the Illumina mate pair and 454 paired reads have templates which are roughly 2.5 kbp in size. The mean size of the megareads, which is essentially the same thing as the mean size of the usable part of the raw PacBIO reads, is 2.47 kbp, with a standard deviation of 3.1 kbp. With these limitations on long range linkage assemblers begin having difficulty building over repeats once these reach about 2 kbp. A rare long PacBIO read may span a much longer read, up to around 6 kbp, but given the coverage, it would be extremely unlikely for there to be two such reads. Accepting linkage based on a single PacBIO read is unwise since that read might be chimeric.
Scaffold mapping onto a BAC control
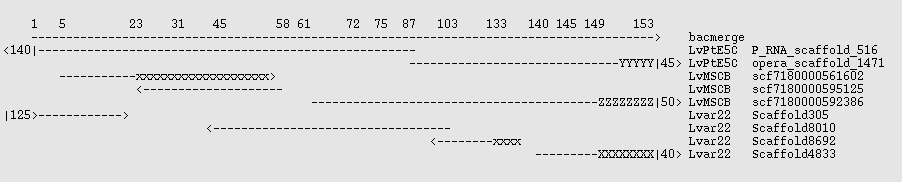
There exists a small set of 27 BAC sequences for this organism which may be used as controls. None of them are completely sequenced, but the pieces are ordered. Two of them, AC146989 and AC131496 overlap. These were extracted and fused with the EMBOSS megamerger program to produce a sequence "bacmerge" 154 kbp long and containing 2200 N characters. Lvar 2.2, LvMSCB, and LvPtE5C were mapped against that with the command
blastn -query /tmp/bacmerge.fasta -subject assembly.fasta -outfmt 6
and the best scaffolds noted. These are shown graphically in the image below, where x,X,Y,Z are mismatches, - is a match, and <N| indicates that the scaffold extends N kbp beyond the end of the reference sequence. x indicates a bad join, which is a problem in two of the assemblies. The 3' ~4.5 kbp of the control is largely composed of repeats having tuples consistent with ~20x copies, and the X,Y,Z sequences in the assemblies each have somewhat different sequences there - none of which match the BAC control well. (Right click on the image and open in a new window if it is distorted.)